Biosynthesis of nucleic acids
New polynucleotides are synthesized in the cell from mononucleotides, with the original (template) strand helping to order the nucleotides of the nascent strand based on regular base pairing. The synthesis of a DNA molecule according to the DNA matrix (template) or the synthesis of RNA according to the RNA matrix is called replication, the synthesis of RNA on the DNA matrix is considered transcription, and the synthesis of DNA according to the RNA matrix is called reverse transcription . During translation, a peptide is synthesized based on the information stored in the RNA nucleotide sequence. Newly formed nucleic acids have the same information contentas their matrix; the new chain is not identical to the original, but complementary and has the opposite polarity.
DNA replication[edit | edit source]
The term replication includes not only the lengthening (elongation) of the chain, but also the initiation (initiation) and termination (termination) of this synthesis.
Biosynthesis in bacteria[edit | edit source]
Bacteria have a number of prerequisites for the processes of storage and expression of genetic information to be known in more detail in them. They divide rapidly and the mechanisms of inheritance are simpler than in nucleated cells. However, the basic principles of these events are the same in both systems. The bacterium Escherichia coli was most often used for the experiments . Meselson and Stahl demonstrated that DNA replication is semiconservative . This means that each of the two strands of the double helix is a matrix for the synthesis of a new strand. The result is two double helices, each a hybrid of one original and one new DNA strand.
Enzymes that catalyze the elongation of the synthesized chain are called DNA-dependent (dependent) DNA polymerases . In vitro experiments have shown that, in addition to the enzyme, DNA elongation requires matrix DNA, simultaneously all four deoxynucleoside triphosphates (dATP, dGTP, dTTP, dCTP), Mg2+ ions and the so-called eye primer, which is a shorter nucleic acid chain (DNA or RNA) with a free 3'-OH end. DNA polymerases are not able to initiate chain synthesis from the mononucleotide itself, de novo .
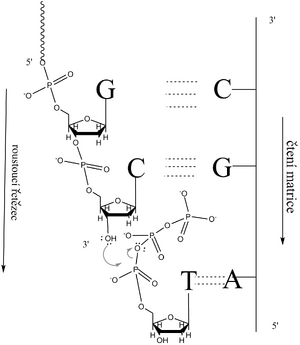
The primer is hybridized to the template strand. According to the first unpaired base of the template strand, located at the 3'-end of the primer, a complementary dNTP is selected. Its α-phosphate is attacked by the nucleophilic oxygen of the terminal 3'-OH of the primer (or growing DNA strand). The transfer of electrons releases the β- and γ-phosphate as pyrophosphate (PPi), while the α-phosphate of the deoxynucleotide is bound to the 3'-OH group of the primer (of the new DNA strand). This is a reverse reaction. However, macroergic PPi is readily cleaved by pyrophosphatase, shifting the balance toward chain synthesis.
The mentioned mechanism implies that the synthesized polynucleotide grows in the direction 5' → 3', while the matrix chain is read in the direction 3' → 5'.
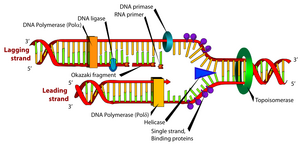
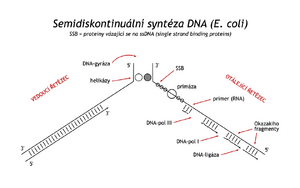
When it was discovered that DNA synthesis proceeds simultaneously along both antiparallel parent strands, which unravel in the process (replication forks), it was necessary to clarify how nature solved DNA synthesis on strands of opposite polarity and how the unraveling of higher DNA structures is ensured. The first of the problems became clear when DNA synthesis was shown to be semi-discontinuous. The so-called the leading strand grows , as already mentioned, continuously and in the 5' → 3' direction, while the synthesis on the antiparallel matrix is more complex. The matrix is suitably coiled and in the direction 5' → 3' a lagging strand is formed on it, in the form of shorter sections, Okazaki fragments. The fragments are then additionally joined into a continuous chain. A primer is needed to initiate the synthesis of each fragment . It is synthesized by a special RNA polymerase called primase . The choice of site for primer synthesis is not random. Primase is part of the primosome protein complex, as is the N protein, recognizing the appropriate sequence at the site of the future primer (the average distance between these sites is 1000–2000 bp). Unlike DNA polymerases, primase is able to initiate chain synthesis from a single nucleotide. It makes RNA shorter than 10 nucleotides. From the 3'-OH end of this primer, DNA-dependent DNA polymerase III synthesizes the Okazaki fragment1000–2000 nucleotides long before reaching the preceding primer. It is then replaced by DNA polymerase I, which in addition to polymerase activity also has 5' → 3' exonuclease activity. The enzyme cancels the previous primer and extends the DNA fragment to the beginning of the previous DNA fragment
The different ability of RNA and DNA polymerases to initiate de novo chain synthesis is related to the fact that DNA polymerases check and possibly correct the pairing of the last nucleotide in the chain before extending it by the next nucleotide. The accuracy of DNA polymerases is an order of magnitude higher compared to the accuracy of RNA polymerases. RNA is probably an older and less precise form of storing and transmitting genetic information than DNA.
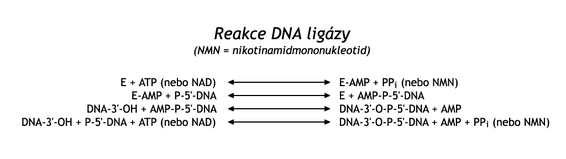
In the process described on the lagging strand, it remains to join the Okazaki fragments in continuous DNA. This reaction is catalyzed by DNA ligase . The enzyme connects the 3'-OH end of one DNA strand with the 5'-phosphate end of another strand through a phosphodiester bond. DNA-ligase is first activated by accepting AMP, originating from ATP or from NAD , on the ε-NH 2 group of its lysine. This complex transiently binds to the 5'-phosphate at the end of the splicing DNA and thereby activates it. Then, the 3'-OH group of the second fragment is attached to this phosphate by a nucleophilic attack. At the same time, AMP is released (see reaction scheme)
DNA ligase has a number of functions. In addition to Okazaki fragments, it connects DNA chains during DNA repair, during "crossing over", and is also used in artificial gene manipulations .
In addition to DNA polymerases, primosome and DNA ligase, other proteins are used in the replication fork. During the simultaneous cleavage of A
TP, the enzyme helicase unravels the double helix . SSB-proteins (single strand binding proteins), i.e. proteins that bind to single-stranded DNA, immediately bind to the released chains to prevent its reassociation at the replication site.
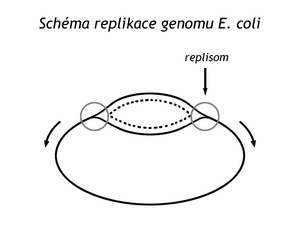
The complex of enzymes at the replication fork is sometimes called a replisome . As the replisome proceeds and unwinds the right-handed double helix, the original DNA must rotate around its axis counterclockwise, creating tension and a positive superhelix. Replication would stop. Topoisomerases solve the problem; DNA gyrase can introduce a negative superhelix, swivelase breaks it. Both enzymes maintain a degree of negative DNA supercoiling, promoting the unfolding of the dsDNA helix.
Iniciation of DNA replication in bacteria[edit | edit source]
Initiation of bacterial DNA replication is a very precisely controlled process. On circular dsDNA (about 4 million bp), there is a single site (ori C) with four repeating sequences of nine base pairs (bp). They are distributed in a 240 bp stretch of DNA. The replication proteins dnaA are attached to them (the name is derived from the gene on which they arise). The DNA naturally coils and other dnaB and dnaC proteins attach to the complex . The dnaB protein has helicase activity, it partially unfolds the double helix, SSB proteins and other helicases bind, and replication is initiated by primer synthesis.
In bacteria and viruses, two replication forks are usually formed at the ori site, which then proceed along the DNA in opposite directions, creating a so-called replication eye
Termination of replication in bacteria[edit | edit source]
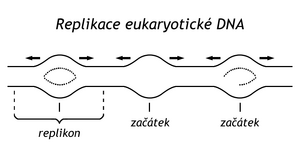
In the circular DNA of bacteria and viruses , replication ends when replisomes meet on the opposite side of the circular DNA. The stretch from the ori site to the replication termination site is called a replicon . In bacteria, this is a circular replicon. However, new circular DNA molecules can be threaded through. Type II topoisomerase can break them apart.
In the case of linear DNA molecules (some viral DNA), a mechanism is required to terminate replication, which resolves incomplete DNA synthesis at the 5'-end of the new strand (the stretch after primer removal). The problem is solved in several ways, e.g. by appropriate end sequences and coiling of the replicated DNA, or by initiating synthesis from both ends of the template dsDNA.
Multiplicity of DNA-polymerases and repair of damanged DNA in bacteria[edit | edit source]
Three DNA-dependent DNA polymerases have been recognized in E. coli :
| Enzyme type | I | II | III |
|---|---|---|---|
| Polymeration 5' → 3' | + | + | + |
| Nuclease activity 3' → 5' | + | + | + |
| Nuclease activity 5' → 3' | + | − | + |
| Number of molecules in a cell | 400 | ? | 10–20 |
| Elongation rate (number of nucleotides/min) | 600 | ? | 10 000 |
| Number of subunits | 1 | ? | 7 |
| Molecular weight | 109 000 | 120 000 | 250 000 |
DNA polymerase I was discovered first because it is the most abundant in the cell. Although it is formed by a single peptide chain with a molecular weight of 109,000, it abounds with three enzyme activities, polymerase and two nuclease. For 3' → 5' exonuclease activity, the 3'-OH group must not be esterified and the cleaved nucleotide must be free, unpaired. The activity is used to modify the wrong end of the primer or to remove the misaligned nucleotide. The 5' → 3' nuclease site is separated from the polymerase center. It cleaves the terminal mono- or oligonucleotide at the 5'-end of the chain (phosphorylated or free), whereby the cleaved nucleotide must be paired with a complementary base in dsDNA. It has been shown that DNA polymerase I is not an intrinsic replication enzyme of E. coli, but a repair enzyme. The structure of DNA is not so constant that it can ensure the known functional stability of genes over time. Spontaneously, by thermal movements, β-glycosidic bonds of purine bases are often broken, cytosine is deaminated to uracil, two adjacent thymines (TT dimers) are covalently bound under the influence of ultraviolet light . DNA polymerase I recognizes the faulty nucleotide, the section in which it is located, the enzyme cleaves it and synthesizes the correct chain ("patch") according to the complementary chain. DNA ligase binds the 3'-OH end of the repaired section to the original strand.
An E. coli mutant with a defect in DNA polymerase I is significantly more sensitive to the action of mutagens, but it divides at the same frequency as normal bacteria . It follows that the proper replication enzyme is a different polymerase. DNA-dependent DNA polymerase III has been shown to have this function. Although the bacterium contains only 20 molecules of the enzyme, it extends the chain fifteen times faster than type I. The holoenzyme of DNA polymerase III consists of at least 7 subunits α, β, γ, δ, θ, ε, τ. In addition to polymerase activity (subj. α), it also has 3' → 5' and 5' → 3' nuclease activity. The function of another enzyme, DNA polymerase II, is not yet known.
Replication of chromosomal DNA in eukaryotes[edit | edit source]
In this chapter, the most important differences between replication in nuclear cells and bacterial DNA replication will be described. In eukaryotic cells, there are four DNA-dependent DNA polymerases (see table), which have the same basic features of polymerase activity as bacterial enzymes: they extend the DNA chain at the 3'-end, dNTPs are the substrate, a matrix and a primer are needed for replication. The main replication enzymes of eukaryotes are DNA polymerases α and δ, each on one parental strand, while DNA polymerase β has a repair function. Enzyme γ replicates mitochondrial DNA.
| Typ enzymu | α | β | γ | δ |
|---|---|---|---|---|
| Polymeration 5' → 3' | + | + | + | + |
| Nuclease activity 3' → 5' | − | − | + | + |
| Relative activity | 80 % | 10–15 % | 2–15 % | ? |
| Number of subunits | several | 1 | 1 | ? |
| Localisation | kernel | kernel | mitochondria | kernel |
| Molecular weight | 140 000 | 40 000 | 150 000 | ? |
| Function | replication | repairs | replication of mtDNA | replication |
On a long eukaryotic DNA molecule, replication begins at many sites simultaneously. A number of replication loops are formed, tens of thousands per DNA molecule of embryos, thousands per DNA molecule of adult animals.
The speed of the replication fork is 4x slower in eukaryotes than in bacteria, so for example the longest chromosome of Drosophila would take 16 days to replicate from one place, it actually takes 3 minutes
Okazaki fragments in nuclear cells are made up of only about 140 nucleotides.
Chromosomal DNA repair is as essential as in bacteria. DNA in each human cell loses 5000 purine bases per day (ternal β-glycosidic bond breaks), 100 cytosines in DNA are spontaneously deaminated to uracil, and ultraviolet light creates covalent dimers from neighboring thymines. Repair mechanisms must reduce the high frequency of introducing genetic errors to a minimum. Patients with the skin disease xeroderma pigmentosum (an autosomal recessive trait) have a defective nuclease that hydrolyzes DNA near thymine dimers. The disease is manifested by hypersensitivity of the skin to sunlight, dryness and atrophy of the skin, keratosis, corneal ulcers, and skin cancer usually develops by the age of 30.
The presence of thymine in DNA and uracil in RNA is also related to genome repairs. Deamination of cytosine to uracil is a potentially mutagenic change. The enzyme uracil-DNA-glycosidase hydrolyzes the glycosidic bond between uracil and deoxyribose. A site without a base is recognized by a specific endonuclease. After splitting the chain, repair DNA polymerase and DNA ligase insert the correct section with cytosine again. If there was uracil instead of thymine in DNA, no enzyme would distinguish between "correct" uracil and deaminated cytosine. The energy spent on the methylation of uracil to thymine is a "tax" for the accuracy in the storage of genetic information. Deamination of cytosine in RNA is not so critical. A particular DNA molecule is only found in one or a few copies in a cell, so a base substitution in DNA is much more serious than a base substitution in one of the huge number of identical RNA molecules.
The repair mechanism also explains how important the double-stranded arrangement of DNA molecules is.
Fate of histones during DNA replication[edit | edit source]
Histones are mainly synthesized during the S-phase of the cell cycle . They then enter the nucleus from the cytoplasm, where they bind to DNA. A small part of histones is probably synthesized during the cell cycle and is probably intended for repair.
Experiments with isotopically labeled amino acids showed that newly synthesized histones form complete new octamers; mixed complexes containing both old and new histones were not found. The new octamers bind to the newly synthesized DNA, they do not exchange with the histones in the non-replicating part of the DNA. While the original histones were found on the leading-strand DNA double helix after replication, the new octamers bind to the lagging-strand DNA.
Transcription[edit | edit source]
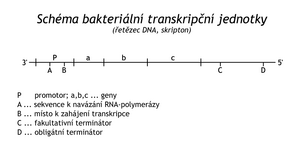
Transcription refers to the synthesis of RNA according to the DNA matrix. Therefore, the enzymes catalyzing this reaction are called DNA-dependent (dependent) RNA-polymerases . They do not require a primer, they start the synthesis on the 1'-OH nucleotide of GTP or ATP. The building blocks in this case are ribonucleoside triphosphates (ATP, GTP, UTP, CTP). The chain elongation mechanism is the same as in DNA synthesis. Thus, during transcription, the RNA grows in the 5' → 3' direction and the DNA template is read in the 3' → 5' direction. The transcribed section of DNA is called a scripton , the newly synthesized RNA is a transcript . It is then modified into functional RNA by several types of reactions . Unlike replication, transcription is asymmetric; only one strand of DNA (negative or anti-coding) is transcribed in a given section. From the point of view of transcription, the second strand of DNA (with the same sequence of nucleotides as in the synthesized RNA) is referred to as positive, coding. However, the opposite strand can be transcribed on the same DNA molecule, but in a different section. Transcription produces several types of RNA, which can be functionally divided into three groups: ribosomal RNA (rRNA) is a structural part of ribosome subunits, transfer RNA (tRNA) transfers activated amino acids to the site of polypeptide synthesis and messenger RNA (mRNA, information RNA, messenger RNA, messenger = messenger) that transfer information about the sequence of amino acids in a peptide from DNA to the site of proteosynthesis on the ribosome.
Transcription in bacteria (E.coli)[edit | edit source]
Different types of RNA are synthesized in the bacterium E.coli . The synthesis of all types of RNA is catalyzed by a single enzyme, DNA-dependent RNA polymerase, consisting of five subunits: 2α, β, β', σ. The α subunit binds the β and β' subunits and is important for the recognition of the scripton start sequence. The β subunit has an active site for the formation of a phosphodiester bond, β is involved in the binding of polymerase to DNA. The σ subunit (also σ-factor) is necessary to recognize the beginning of the scripton (the so-called promoter ).
Initiation of bacterial transcription[edit | edit source]
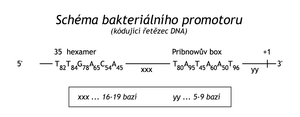
Transcription is initiated at a stretch of DNA called a promoter . The nucleotide sequence in bacterial promoters is analogous in different transcription units. The first transcribed nucleotide is numbered +1. Around the 10th nucleotide against the direction of transcription is the so-called Pribnow box , which is a sequence that does not differ much from the 5' TATAAT 3' sequence. The closer the Pribnow box is to this sequence, the more often the promoter is recognized by RNA-polymerase and the more often transcription is initiated there ("strong" promoter). Around the 35th nucleotide there is usually another general sequence, mainly 5' TTGACA 3', which also contributes to the strength of the promoter.
RNA-polymerase binds randomly and weakly to any site on the DNA, moves along the matrix for a short distance and is released again. If it finds a promoter this way, it binds more tightly. The s-factor contributes to the specificity of binding to the promoter. The polymerase unfolds a short section of the DNA double helix (about 17 bp) and starts transcription at the +1 nucleotide. Unlike ONA polymerase, RNA polymerases do not require an oligo- or polynucleotide primer (loop), they start the synthesis of a new chain either from adenosine triphosphate or guanosine triphosphate.
Elongation of the synthesized RNA strand[edit | edit source]
After the binding of the first few nucleotides of the new RNA chain, the σ-factor is released from the polymerase, the polymerase proceeds along the matrix (anticoding DNA strand) and synthesizes RNA (transcript). About 10–12 nucleotides of the newly synthesized RNA remain bound (hybridized) at the site of transcription, the extended RNA chain is released after this section, unfolds. The transcribed section of DNA coils again into a double helix. Unlike DNA polymerases, RNA polymerases do not have nuclease activity, they do not correct misplaced nucleotides. Transcription is not as accurate as replication (DNA synthesis).
Termination of transcription in bacteria[edit | edit source]
The polymerase proceeds to a section of DNA called a terminator . Sequentially, it is a G- and C-rich inverted repeat interrupted by a unique sequence. The terminator transcript creates a looped hairpin. This is followed by a 4-8 dA oligosequence, transcribed into the corresponding oligo-U section of the transcript. The DNA-RNA hybrid is more labile in the section with repeating A=U pairs than the RNA-RNA hybrid in the hairpin with frequent G=C pairs. Therefore, the transcript is released. Transcription is always terminated at this terminator, which is why it is called an obligate terminator . Obligatory terminators are often preceded by so-called facultative terminators . They do not contain the oligosequence d AAAA.., a regulatory protein, ρ-factor (rhó) is needed to terminate transcription, which binds to the RNA hairpin and helps release the transcript. After stopping the RNA-polymerase, releasing the finished RNA and releasing the polymerase from the DNA, the template DNA is completely coiled again into a double helix. Scripton is regulated as a transcriptional unit, in this sense it is called an operon in bacteria . It is characteristic of a bacterial operon that tandemly arranged genes are transcribed into a single large transcript. In the case of structural genes (encoding polypeptides), the transcript is a so-called polycistronic (multigene) mRNA. Ribosomes then synthesize individual peptides on this tandem of information. Unlike bacteria, there is no mechanism in nuclear cells to separate this information, so in eukaryotes functional peptides are often separated only as part of post-translational modifications of a large protein precursor. For example, several peptide hormones are formed from one large protein
Inhibitors of bacterial transcription[edit | edit source]
Some substances interfere with a certain phase of transcription and thus inhibit it. The antibiotic rifampicin binds to the β-subunit of RNA polymerase; the enzyme can bind to DNA, but does not initiate transcription. Streptolygidine stops the elongation of the transcript by binding to the 13-subunit of the polymerase. The antibiotic actinomycin D is often used in experiments . It inhibits not only bacterial but also eukaryotic transcription; its phenaxozone ring intercalates (inserts) between DNA bases and each of its peptide chains forms a strong hydrogen bond with the -NH of guanine. The polymerase is stopped by the antibiotic. The acidic glycosaminoglycan heparin binds to the strongly basic β'-subunit of RNA polymerase and prevents the polymerase from binding to DNA.
Post-transcriptional modifications (maturation, processing) of bacterial RNA[edit | edit source]
Newly synthesized RNAs are edited into functional molecules. In prokaryotes, only so-called "permanent" RNAs, i.e. rRNA and tRNA, are subject to this maturation, while mRNAs with a half-life of several minutes are synthesized in a functional form. Even during transcription, ribosomes are attached to incomplete mRNA in bacteria, translation takes place simultaneously with transcription, as well as degradation of mRNA after the last ribosome. In nucleated cells, transcription and translation are temporally and spatially separated, while mRNA undergoes complicated maturation processes. Transcripts of permanent bacterial RNAs contain several functional RNA molecules that are removed from these precursors by the action of specific exo- and endonucleases. There are about 10 species in E. coli , a few of them are listed in the table:
| Nukleázy podílející se v E.coli na posttraskripčních úpravách RNA | ||
|---|---|---|
| Enzyme | It splits | Specified |
| R nase P | tRNA 5’-end | endo- |
| R nase D | tRNA 3’-end | exo- |
| R nase T | tRNA CCA | exo- |
| R nase III | rRNA (and phage mRNA) | endo- |
| R nase H | RNA-DNA hybrid | endo- |
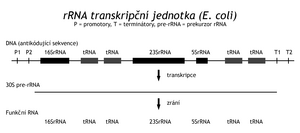
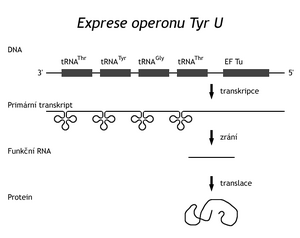
These nucleases recognize the site of action by the conformation of the substrate, not by the sequence of the cleaved nucleic acid. The rRNA genes are located in a cluster with the genes of several tRNAs. Transcription of the operon creates a 30S rRNA precursor, which is then cleaved by RNase III into sections containing functional molecules. Other enzymes then modify these RNAs into their final functional form. Other transfer RNAs are also synthesized in clusters. In the genome of "E.coli" there is a so-called TyrU-operon, containing genes for two different tRNAs Thr , tRNA Tyr , tRNA Gly and a gene for an elongation factor needed during translation. In another TyrT operon, there are two identical Tyr tRNA genesand the gene for protamine (protein P). Other tRNAs are transcribed from monogenic transcription units. Transcription of the TyrU-operon results in an RNA precursor that is first cleaved by RNase P (endonuclease) at the definitive 5' ends of the tRNA. In addition to the peptide chain, RNase P also contains RNA (375 bases). Exonuclease modifications of the 3'-ends of the tRNA probably follow. RNase D cleaves them up to the CCA sequence, recognizing the 3'-end by conformation. The transcript may not contain the CCA sequence if it is not encoded in a gene. In bacteria, we distinguish 2 types of tRNA. Type 1 contains a 3'-terminal CCA sequence in the gene, type II does not. In the second case, the missing part of the -CCA sequence is added post-transcriptionally by the action of tRNA nucleotidyl transferase. It is another type of vision (and repair) mechanism.
Transcription in animal cells[edit | edit source]
Analogous types of RNA are synthesized in nucleated cells as in bacteria. Additionally, in eukaryotes there is one small rRNA (5SrRNA), a heterogeneous RNA that contains precursors for mRNA (pre-mRNA), small nuclear (snRNA) and nucleolar RNA (snoRNA), and mitochondrial RNA (mtRNA). In addition to mitochondrial RNA, transcripts are produced in the nucleus and then, mostly in the form of ribonucleoprotein, undergo modification and eventually move to the cytoplasm. RNA synthesis in eukaryotes is catalyzed by four types of DNA-dependent RNA polymerases (see table). Each of them is composed of 10−15 subunits (protomers).
| Basic types of eukaryotic RNA | ||||
|---|---|---|---|---|
| Precursor | Functional type | Molecular weight | Localisation of the cell | Representation (%) |
| 45S pre-rRNA | 4,5x106 | nucleolus | 1 | |
| 35S | ||||
| 28S | 5,8S rRNA | ribosomal subsequences 60S | ||
| 28S rRNA | 1,7x106 | 53 | ||
| 23S | 18S rRNA | 0,7x106 | ribosomal subsequences 40S | 24 |
| 5S rRNA | 4,2x104 | ribosomal subsequences 60S | 1 | |
| pre-tRNA | 4S tRNA | 2,3–3x104 | cytoplasm | 12 |
| hn-RNA | ||||
| pre-mRNA | 6–30S mRNA | 3x104 (2x10 cytoplasm) | polysomes | 3 |
| snRNA | ||||
| snoRNA | ||||
| mtRNA | mitochondria | |||
| Typy eukaryotických RNA-polymeráz | |||
|---|---|---|---|
| RNA-polymeráza | Lokalizace | Citlivost k inhibitorům | Produkt |
| I | nucleolus | resistance of α-amanitin | 45S rRNA |
| II | nucleus | very sensitive of α-amanitin | hnRNA, (pre-mRNA), snRNA |
| III | nucleus | moderately sensitive of α-amanitin | tRNA, 5S rRNA, snRNA |
| mitochondr. | mitochondria | sensitive of rifampicin | mtRNA |
Each RNA polymerase has a characteristic type of promoter. The polymerase II promoter is best known so far. Its structure is analogous to a prokaryotic promoter.
Initiation of eucaryotic transcription[edit | edit source]
The long DNA molecule in the chromosome is intricately coiled and bound with proteins. The section to be overwritten must be made available. The mechanisms of DNA unfolding are being studied. Several regulatory proteins suppress deacetylase activity so that acetylation of histones by acetyltransferase predominates. The nucleosome chain is transiently unfolded, after the local loss of nucleosome histones, the DNA is converted into a supercoiled conformation. The energy of this conformation is used to regionally unwind the DNA helix, allowing transcription to begin. After the dephosphorylation of the regulatory proteins, deacetylation again prevails, enabling the restoration of histone octamers, nucleosomes and DNA coiling. In the 5S rRNA gene, the promoter for RNA-polymerase III is located only after +1 nucleotide in the gene. Apparently, a factor is required for polymerase binding, Elongation of animal RNAs takes place by the mechanism described in bacteria. Termination in eukaryotes has not yet been studied in detail.
Inhibitors of eucaryotic transcription[edit | edit source]
Actinomycin D inhibits transcription in a nucleated cell as well as in a bacterium. Another proof for the endosymbiotic theory of the origin of mitochondria is the analogy in the sensitivity of mitochondrial RNA-polymerase and bacterial RNA-polymerase to inhibitors (e.g. rifampicin ). α-amanitin is a peptide produced by some higher fungi, especially the poisonous green toadstool (Amacita phalloides). The toxin binds to RNA polymerase II and preferentially stops mRNA synthesis. Liver cells are the most affected.
Post-transcriptional modifications (processing, maturation) of eukaryotic RNA[edit | edit source]
rRNA edits[edit | edit source]
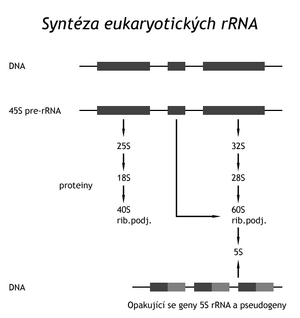
All three major types of RNA (rRNA, tRNA and mRNA) have been shown to be post-transcriptionally modified in eukaryotes. Three of the functional rRNAs, ie 18S rRNA, 5.8S rRNA and 28S rRNA are contained in the primary transcript of the 45S pre-rRNA. The 45S pre-rRNA is metabolically more stable than the prokaryotic 30S pre-rRNA transcript. It is enzymatically cleaved and modified into three functional rRNAs. About 100 nucleotides are methylated to 2'-OH and more than 100 uridines are isomerized to pseudouridine. Unlike prokaryotes, the rRNA gene does not contain tRNA genes. Eukaryotic cells synthesize yet another ribosomal RNA – 5S rRNA, on a repeating gene, always accompanied by a non-functional pseudogene. This molecule is probably not post-transcriptionally modified.
tRNA edits[edit | edit source]
There are several hundred to thousands of tRNA genes in eukaryotic cells. Thus, the genes for a particular tRNA are repeated many times, these are repeats scattered throughout the genome. The primary transcript is shortened by end-editing. In eukaryotes, tRNA splicing is also known , which is the cleavage of a certain internal section ( intron ) of pre-RNA. Introns of eukaryotic pre-tRNAs do not contain characteristic flanking sequences. Cleavage sites are recognized by a specific endonuclease based on the tertiary structure of the pre-tRNA. The two resulting fragments are connected by RNA-ligase while cleaving ATP. Considerably different splicing mechanisms have been discovered for eukaryotic pre-mRNAs.
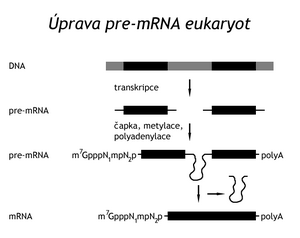
mRNA edits[edit | edit source]
Transcription of a structural gene produces pre-mRNA (their mixture is sometimes referred to as heterogeneous nuclear RNA, hnRNA ). Proteins bind to this primary transcript. In doing so, it is subjected to four types of post-transcriptional modifications:
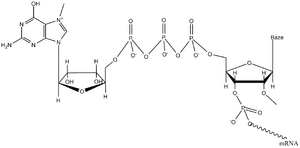
Cap connection (capping). Also in eukaryotes, transcription starts on ATP or GTP. The first modification of the emerging pre-mRNA is the attachment of a 7-methylguanine nucleotide to the 5'-triphosphate end of the still unfinished chain. First, one phosphate is cleaved from it, and the diphosphate end then attacks the α-phosphorus of free GTP. This creates an unusual 5'–5' anhydride bond between RNA nucleotides. N7 of guanine and 2'-OH of the first and second nucleotides tend to be methylated. The cap protects the 5'-end of the molecule from exonucleases, and after binding the CBP-protein (cap binding protein), it is recognized by the translation initiation factor elF-3, which enables the correct binding of the mRNA to the ribosome
Internal methylation. Inside the chain, mRNA is methylated mainly at adenosine in position 6 (m 6 A). The meaning of this modification has not been clarified
Polyadenylation . Most eukaryotic mRNAs have a polyadenylated sequence at the end. This modification is initiated by a specific endonuclease that recognizes the sequence AAUAAA near the 3'-end of the transcript. Another enzyme , polyA-polymerase , sequentially adds about 250 adenine nucleotides to the mRNA. Several molecules of a special protein are then bound to them. Polyadenylation is inhibited by 3'-deoxyadenosine ( cordycepin ). Also, the meaning of this modification is not clear
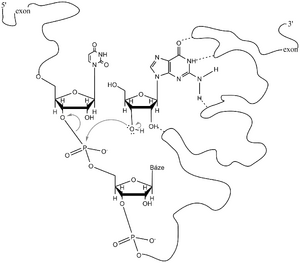
Splicing . Unlike prokaryotic genes, eukaryotic structural genes are interrupted by sections, so-called introns , which are not part of functional mRNA. The remaining sections, which after transcription are joined into a chain of mature RNA, are called exons . The eukaryotic cell is equipped with a mechanism that brings together the 3'-end of the proximal and the 5'-end of the following exon and then excises the intron and joins the two exons. The process is called splicing. Each intron in all eukaryotes and all pre-mRNAs begins with the sequence GUa and ends with AG. 20–50 nucleotides towards the 5'-end from the 3'-end of the intron is an important sequence called a branch site, which contains adenine. Splicing begins with cleavage of the phosphodiester bond between the 3'-end of the proximal exon and the 5'-end of the intron. The phosphate of this bond is transferred to the 2'-OH of the aforementioned adenine nucleotide at the branch point. It is an energy-efficient transesterification, during which a nucleotide loop (loop) is formed. The released 3'-OH at the end of the proximal exon attacks the phosphorus atom between the 3'-end of the intron and the 5'-end of the distal exon. In this way, both exons are covalently joined and the so-called lasso-like RNA containing the intron sequence is released
One of the important features of pre-mRNA splicing is the fact that it does not involve hydrolysis, but transesterification, so the whole process takes place at approximately the same energy level, unlike pre-tRNA splicing. It is, of course, important that the 3'-end of one exon approaches the 5'-end of the following exon several hundred nucleotides away and joins it with the precision of a single nucleotide. In mammals, this problem is solved by a ribonucleotide complex (60S), called the spliceosome . It contains several small nuclear RNAs (snRNAs). U1 snRNA has a sequence complementary to the site at the 5'-end of the intron, U2 snRNA recognizes the branching site. U5 snRNA binds to the 3'-end of the intron. In addition to proteins, the spliceosome also contains U1 and U6 snRNA.. In the protozoan Tetrahymena, an intron is excised from the 26S rRNA precursor without the involvement of proteins. Thomas Cech (originally Czech) and Thomas Altman (Nobel Prize winners) and collaborators discovered that an RNA molecule can create an active enzyme site for itself and catalyze its splicing, i.e. self- splicing. The reaction requires a cofactor, which can be any guanine ribonucleotide (GTP, GDP, GMP, but also the nucleoside guanosine). The edited RNA conforms to form a specific enzyme site into which the guanine cofactor binds. The 5'-end of the intron is transesterified to its 3'-OH. The released hydroxyl group of the 3'-end of the proximal exon then attacks the 5'-phosphate of the distal exon, releasing the intron as a linear molecule. Catalysis is strictly seterospecific as we know it in protein enzymes. After several modifications (including shortening by 19 nucleotides), a circular RNA known as L19 RNA is formed from the released linear intron. It has an active site that binds the guanine cofactor and has special nuclease and polymerase activity, cleaving and elongating oligonucleotides. That is why it is called a ribozyme. Scientists believe that early in the evolution of life, RNA could have replicated itself without the involvement of proteins. Another type of self-splicing (type II) was found in the mitochondria of molds and fungi, which is very similar to the described self-splicing I (Tetrahymena). The only difference is that type II does not require a gaunin cofactor because the 5'-end of the intron is transesterified to the 2'-OH of the branch site adenine nucleotide. The intron is released as a lasso circular RNA. Type II self-splicing is thus a kind of developmental intermediate stage between type I and the spliceosome type of higher eukaryotes
Bibliography[edit | edit source]
- ŠTÍPEK, Stanislav. Stručná biochemie. 1. edition. Medprint, 1998. ISBN 80-902036-2-0.