Respiratory Chain and ATP Generation (FBLT)
Content of subsection
- Introduction to the issue of the respiratory chain and ATP production
- Mitochondrial electron transport chain
Introduction to the issue of the respiratory chain and ATP formation
Oxidation is the loss of electrons or the increase in the oxidation number of the respective element. The removed electrons must end up on another element, which is why the oxidation of one substance is always associated with the reduction of another substance. The word "oxidation" comes from the word "oxide", which refers to a compound containing oxygen. Oxygen is not absolutely necessary for oxidation to occur.
Oxygen is an oxidizing agent. It has a high tendency to accept electrons. The affinity of an element for electrons is expressed by the value of its electronegativity. The electronegativity of oxygen is one of the highest among the elements, only fluorine has a higher electronegativity. The transfer of electrons to oxygen, including the formation of bonds with oxygen, is a thermodynamically advantageous process, i.e. releases energy.
Organisms living on Earth have found a way to use energy by transferring electrons from less electronegative elements to oxygen. Our cells oxidize organic compounds contained in food to CO2' and H2O' for "oxygen consumption" and "energy production". This process is also sometimes called nutrient burning. Unlike combustion, the transfer of electrons from organic molecules to oxygen in our cells is divided into many steps. Most of the released energy is not converted into heat and light, but is stored in the form of a chemical potential. For didactic reasons, we can divide oxidative metabolism into two phases:
- Substrate oxidation associated with reduction of enzyme cofactors;
- Reoxidation of reduced cofactors by oxygen.
Breakdown of glucose
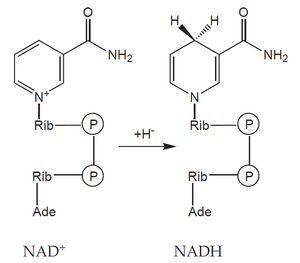
An example is glucose. Its six-carbon molecule (C6H12O6) is gradually oxidized into two molecules in the process of glycolysis and the pyruvate dehydrogenase reaction acetic acids (in the form of acetyl-coenzyme A) and two molecules of CO2. During this reaction, four molecules of the coenzyme NAD+ (nicotinamide adenine dinucleotide) are reduced to NADH, each accepting two electrons (often indicated as acceptance of the hydride anion H< sup>–).
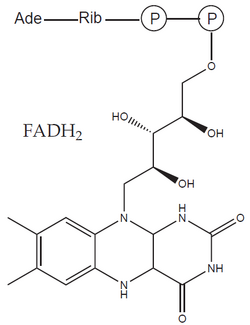
The formed acetyl-CoA enters the citrate cycle and is further oxidized to two molecules of CO2 and four molecules of reduced cofactors (three NADH and one FADH2 – flavin adenine dinucleotide). As a result, during the oxidation of glucose, 12 reduced cofactors'' and six molecules of CO2 are formed. No molecular oxygen (O2) was consumed in this process – the missing oxygen atoms were provided by water molecules. In it, oxygen is already present in a reduced form (O–II), which does not participate in electron transfer.
In order for the metabolic pathway to function, the reduced cofactors must be retroactively 'reoxidized. The process of reoxidation occurs most often in mitochondria, where reduced cofactors transfer the obtained electrons to oxygen (reduce it). The reduction of oxygen then leads to the release of a significant amount of energy.
Mitochondrial electron transport chain
Mitochondria are bound to the cytoskeleton in the cell. According to Lynn Margulis's endosymbiotic theory, they evolved from aerobic bacteria that were previously engulfed by phagocytic archaebacteria. Part of their genome moved into the nucleus of the "host" cell and thus became dependent on it, but the rest of the genome remained in the mitochondria. In addition to their own genes, they also have their own proteosynthetic apparatus showing the characteristics of prokaryote (70S ribosomes), and are therefore referred to as semiautonomous organelles.
Mitochondrial Outer Membrane
They are separated from the external environment by a membrane similar to the endoplasmic reticulum. This membrane is relatively well permeable to most substances with a lower molecular weight, but prevents the entry of proteins and other macromolecules. At the same time it contains:
- enzymes of fatty acid and phospholipid metabolism;
- the so-called TOM complex (translocase of the outer membrane) transferring proteins from the cytoplasm to the intermembrane space.
The intermembrane space has a composition similar to the cytosol (ie, the protein content in it is low compared to the matrix) and contains, for example, cytochrome c and proapoptotic proteins.
Inner Mitochondrial Membrane
Membrane highly selective, polar molecules almost do not pass through it (with the exception of a few that have their own transporters). The inner membrane contains:
- phospholipid cardiolipin;
- respiratory chain enzymes;
- so-called TIM complex (translocase of the inner membrane) transporting certain proteins.
It extends towards the matrix in the form of various protrusions, most often cristae and tubules.
Mitochondrial matrix
It has the form of a dense protein gel containing enzymes of many metabolic pathways (Krebs cycle, β-oxidation of MK, ornithine cycle and others). Furthermore, mDNA (and the corresponding tRNA and mRNA), ribosomes or inorganic ions (such as Ca2+ ).
Electron transport chain
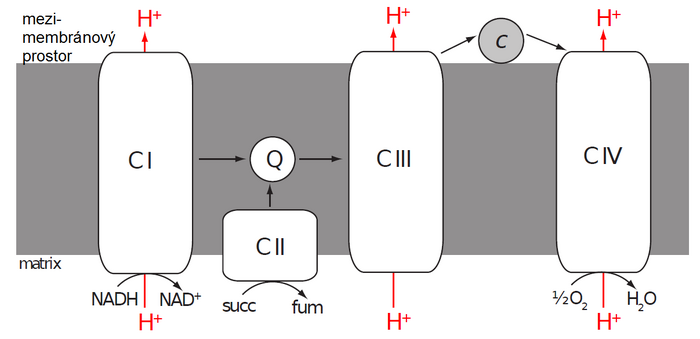
"Reduced cofactors" coming from the "cytoplasm" (via a special transport system), as well as from reactions taking place in the matrix, are reoxidized on the inner mitochondrial membrane by a set of enzymes called the electron transport chain ("ETC - electron transfer chain). It consists of four enzyme complexes referred to as Complex I–IV.
Complex I (NADH dehydrogenase)
- Complex I catalyzes the oxidation of NADH to NAD+ and at the same time the transfer of two electrons to coenzyme Q (CoQ) also called ubiquinone . The exact structure of mitochondrial Complex I is not completely known, but we know that it contains over 40 subunits, one FMN molecule and several iron atoms in complexes with sulfur (FeS clusters). Electrons released from NADH bind to FMN and subsequently jump from one FeS cluster to another until they reach ubiquinone and reduce it to ubiquinol.
- Ubichinol (CoQH2 or UQH2) is therefore a reduced form of ubiquinone (CoQ or UQ). The term coenzyme Q includes both forms. It is an extremely hydrophobic molecule, due to its long isoprenoid side chain, by which it is attached to the nonpolar core of the inner mitochondrial membrane. Here, CoQ functions as a mobile electron carrier from Complex I and II (or other enzymes) to Complex III.
Complex II
- Complex II catalyzes the oxidation of succinate to fumarate' and is an integral part of the citrate cycle (succinate dehydrogenase). Electrons obtained by oxidation are first transported to FAD bound in the enzyme and subsequently via a chain of three FeS clusters and cytochrome b to ubiquinone.
Complex III (cytochrome c reductase)
- Complex III receives electrons from reduced CoQ' and passes them (via two cytochromes and FeS cluster) to another mobile electron carrier - cytochrome c. Cytochrome c is a small hemoprotein attached to the outer surface of the inner mitochondrial membrane. Unlike previous enzymes, one molecule of cytochrome c can transfer only one electron. There is a reduction of iron' heme from ferri' (III) to ferro' (II) form. Thus, the electrons from ubiquinol are transferred one by one through a complex process called the Q-cycle.
Complex IV (cytochrome c oxidase)
- Complex IV, the last member of the ETC, takes the electrons from the "reduced cytochrome c" and passes them through two cytochromes and three copper atoms to the "final recipient" (acceptor), which is "oxygen" '.
Redox Potential
In order for the mitochondrial electron transport chain (ETC) to function as described, there must be a force that "pushes" electrons from NADH to molecular oxygen through it. In the case of burning wood, we talked about the electronegativity of elemental oxygen. A related measure of electron affinity is the redox potential.
In the subsection What drives our cells, we described the electrode potential created by immersing a rod of pure metal in a solution of its ions (i.e. its oxidized form). If we separate the two half-reactions present in each redox reaction (reduction and oxidation), we can define standard electrode potentials for them under standard conditions. Depending on the direction of these reactions, we call them standard oxidation and standard reduction potential. Collectively, we speak of redox potential (usually describing a reaction in the direction of its typical course).
The flow of electrons in the right direction (ie from NADH to oxygen via complexes and mobile electron carriers) in the ETC can be explained by the fact that the redox (more specifically, reduction) potentials of all "stops along the way" gradually increase. This means that as you progress through the chain, its individual links become easier and easier to reduce. The oxygen at the end of the chain is by far the easiest to reduce - it is a very good oxidizing agent.
Energy
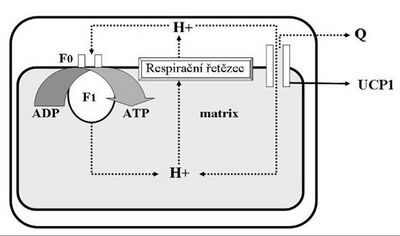
The advantage of using oxygen as the final electron acceptor' is the amount of energy that is subsequently made available. Also, the large increase in reduction potential between NADH (or FADH2) and oxygen and the corresponding change in free energy (ΔG) is not wasted. It is used to pump protons (H+) from the matrix of the mitochondria to the intermembrane space.
The flow of electrons through the Complexes I, III and IV' is associated with the pumping of a certain amount of protons to one pair of electrons. Complex II does not transfer any protons. Since the inner mitochondrial membrane is highly impermeable to protons, a proton gradient occurs on it (the amount of protons is greater in the intermembrane space than in the matrix). A higher concentration of protons means a lower pH and a positive electrical potential. The intermembrane space therefore has a more acidic environment and is positively charged with respect to the matrix. The membrane potential of a mitochondrion is usually expressed as a voltage.
In the mitochondrion, the energy of the proton gradient on the inner membrane is used to produce another kind of energy – 'chemical energy stored in ATP molecules.
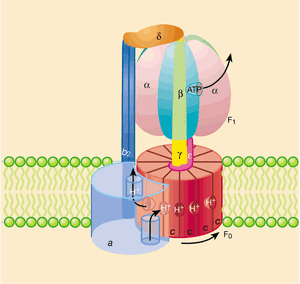
ATP synthesis is catalyzed by the enzyme F1·F0-ATP synthase'. The F0 subunit forms a channel across the mitochondrial inner membrane, allowing the return of protons from the intermembrane space back into the matrix. The flow of protons down the electrochemical gradient is used to rotate part of the enzyme. The rotation is then transmitted to the central axis (stalk) of the F1 subunit of the enzyme, which presses on the outer subunits. These are held immobile by a connection with the peripheral stalk (axis), and thus the phosphorylation of ADP to ATP is driven on them. The F1 subunit has three ATP synthesis sites, so one complete turn will allow the formation of three ATP molecules.
A specific transporter (ANT – adenine nucleotide translocator) subsequently transfers the newly synthesized ATP molecules in exchange for ADP out of the matrix and into the cytoplasm.
Stoichiometry
All the processes described above serve primarily to obtain energy from the substrate for useful work. It is therefore understandable that we are interested in how much energy mitochondria can extract from, for example, glucose or palmitic acid molecules.
In the previous sections, we dealt with the amount of electrons (in the form of reduced cofactors) taken during glucose metabolism. Similarly, we can find out their number for other substrates. So the question of mitochondrial stoichiometry is: how many molecules of ATP can we make in the ETC for a certain number of electrons transferred? We can further divide this question into two sub-questions:
- How many protons are transported across the membrane for one pair of electrons?' We transfer ten protons for two electrons. Complexes III and IV together transport six protons, and complex I will transfer approximately four protons per electron pair.
- How many protons must be moved back into the matrix to make one molecule of ATP? Here the answer is a bit more complicated. According to current models of F1·F0-ATP synthase, the H+/ATP ratio is approximately 4.33. This means that thirteen protons need to be transferred to produce three ATP molecules – ten via F1·F0-ATP synthase and the other three protons are used for the ANT translocator to import ADP and phosphate and to export ATP.
However, the mentioned numbers can only be achieved under optimal conditions, when all components of the ETC work flawlessly and the inner mitochondrial membrane is completely impermeable to protons. Of course, these conditions are not usual.
Uncoupling
The energy transfer mechanism between the ETC and ATP synthesis can be uncoupled if the reverse flow of protons from the intermembrane space to the matrix is allowed. This ineffective loss of the proton gradient transforms the energy stored in it into its less useful form - heat.
Heat is generally much less useful than, for example, ATP, but there are situations where the ability to produce heat can save life - for example, when an organism is exposed to cold temperatures.
Muscle shivering is a well-known heat-generating mechanism used by humans and other mammals. This process produces heat through inefficient energy transfer during muscle contraction. Newborns and many animals use a different method - uncoupling of the mitochondrial chain (uncoupling) = non-shivering thermogenesis.
Uncoupling the respiratory chain to produce heat takes place mainly in a special tissue called brown adipose tissue.
Brown fat:
- is colored like this because it contains many mitochondria,
- its role is simply to "waste" energy.
White fat:
- stores energy.
Mitochondria of brown adipose tissue contain a special protein that forms a channel in the inner mitochondrial membrane and thus enables the transfer of electrons – UCP-1 (uncoupling protein-1, thermogenin). When brown adipose tissue is activated by noradrenaline (via β3-adrenergic receptors), it hydrolyzes its triacylglycerols and the released fatty acids provide energy for ETC and simultaneously activate UCP-1.
There are also other subtypes of UCP (UCP-2 to UCP-5) expressed in other tissues, but their function is still not fully understood.