Polymerase Chain Reaction (detailed)
For molecular examination, it is often necessary to obtain a relatively large amount of a certain section of DNA. Until 1983, there was only one way to multiply a stretch of DNA if it could not be directly isolated in sufficient quantity from the collected material: to introduce it into a bacterial plasmid and clone it. Today, the same is done on an ever-increasing scale entirely in vitro using the polymerase chain reaction PCR. The reaction takes place in a thermocycler, this device changes the temperature at the required intervals.
PCR was invented by Kary Mullis in 1983[1], while he was driving a car in the mountains of California thinking about modifications of sequencing techniques using dideoxynucleotides. Ten years later, he won the Nobel Prize for his discovery. The invention of PCR was truly a revolution for molecular biologists, and nowadays, thanks to its sensitivity, specificity and speed, PCR is probably the most used technique in this field.
Principle[edit | edit source]
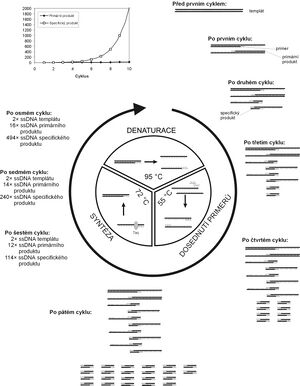
The basic principle of PCR is repeated controlled denaturation of double-stranded DNA and subsequent renaturation of isolated chains with specific oligonucleotides that are in excess in the reaction mixture. These oligonucleotides subsequently serve as primers for the synthesis of a new DNA chain. DNA amplification occurs in repetitive cycles that have three steps:
- 1. Denaturation.
- By heating the DNA to a temperature of around 95 °C, the hydrogen bonds between the DNA strands are broken, splitting the dsDNA into ssDNA.
- 2. Hybridization (anchoring of primers).
- In the literature, this phase is often referred to by the English term annealing (to anneal= to anneal, to temper metal or glass, i.e. to cool them sharply after being heated). It takes place most often at temperatures around 50-60 °C. Single-stranded DNA molecules renature after cooling. If specific oligonucleotides are present in excess in the mixture, they will hybridize to their complementary sequence faster than long single-stranded molecules whose concentration is much lower. The temperature at which the hybridization takes place is critical to the PCR result and must be appropriately set for the primer pair used. At too low a temperature, primers can anneal even to sequences that are only partially complementary, and a non-specific product is thus formed. If the temperature is too high, the primers will not hybridize enough and the product will not be formed in sufficient quantity.
- 3. Elongation, extension, synthetic phase.
- The synthesis of new chains takes place at a temperature of 65–75 °C. Oligonucleotides that have attached to single-stranded DNA (template) in the previous step serve as primers for DNA polymerase in this step. The synthesis of a new chain complementary to the template begins from their 3'-end.
After the first cycle of PCR, the number of DNA strands in the mixture doubles. In the next cycle, the newly formed chains can also serve as a template for the polymerase, so that twice the amount of product is synthesized. As the cycles repeat, the amount of chains created will increase exponentially.
New DNA molecules begin to synthesize from the primer. In the first cycle, when a long molecule of the original DNA serves as a template, chains will be formed which, although shorter than the template, always exceed the section defined by both primers on one side (primary product). If the primary product created in this way serves as a template in subsequent cycles, then a section of DNA with a length precisely bounded by the position of the two primers used will be created according to it (synthesis will start from one primer and end at the location of the other, as this is where the primary product ends). It is precisely these specific products with a precisely defined length that will increase exponentially during PCR, while longer chains will only increase linearly. After 30 cycles of PCR, theoretically about 109 are formed× more specific product than the other DNA sections, the proportion of which is thus practically negligible in the resulting mixture.
The actual yield of PCR tends to be considerably lower than the mentioned 109 copies per one DNA molecule after 30 cycles. This is due to the gradual depletion of reaction components during amplification. After a certain number of cycles, the product concentration reaches a plateau and practically does not increase further.
Components of the reaction mixture[edit | edit source]
Template[edit | edit source]
The requirements for template DNA for PCR are not high, a very small amount of nucleic acid is often sufficient for the reaction. However, it is necessary that the sample is not contaminated with any other DNA that could get into it from insufficiently clean tools, the worker's hands, etc. Even a small admixture of contaminating DNA could multiply during the reaction to such an extent that a detectable amount of product would be produced and the result of the examination would be distorted. In addition, the template DNA sample must not contain substances that would inhibit DNA polymerase. These are often common reagents used in the isolation, purification and processing of DNA (phenol, proteinase K, higher concentrations of salts, heparin, boric acid, ethanol, EDTA etc.).
Primers[edit | edit source]
As a rule, oligonucleotides with a length of 17 to 28 nucleotides are used as primers. In order for the reaction to proceed well, primers should meet several requirements:
- the primers must be specific for the amplified sequence and should be completely complementary to the region to which they are to antagonize,
- primers should not be mutually complementary, to each other , otherwise they themselves will start to act as templates and their dimers will form during PCR (this applies especially to their 3-'ends),
- the primer should not contain internally complementary sequences to avoid internal hybridization (formation of "loops"),
- the distribution of AT and CG pairs should be as even, as possible , especially the 3'-end should not be too rich in CG,
- the temperatures at which both used primers touch should not differ significantly,
- the melting temperature of the primer is at least 50 °C,
- similar melting point of both primers.
It is obvious that two primers are required for the PCR reaction. The primer that starts at the 5'-end of the gene chain and elongates in the direction of transcription during PCR is called the coding, forward primer or upstream primer. The second primer is then labeled anticoding, reverse or downstream.
Polymerase[edit | edit source]
In theory, any DNA polymerase could be used for PCR. However, due to the high temperature in the denaturation phase, at which the enzyme would be denatured, it would be necessary to add the polymerase in each cycle. Therefore, thermostable DNA polymerases, , originally isolated from bacteria living in hot springs, are used for PCR today. The most important representative is the so-called Taq polymerasa from the bacterium Thermus aquaticus. Its temperature optimum is around 75 °C, when it can add about 150 bases per second to the primer. It is not active at temperatures above 90 °C, but it sufficiently resists denaturation.
Today, polymerases for PCR are produced by recombination technologies. In addition to the mentioned Taq polymerase, several other enzymes are used, which, for example, have higher activity, are more thermostable, can synthesize longer chains (Taq polymerase "falls off" after a few hundred added bases from the chain) or have 3'-exonuclease activity, which allows them to repair misplaced nucleotide (Taq polymerase makes one mistake per 10 to 20 thousand inserted bases).
Basic reaction solution[edit | edit source]
The reaction mixture must also contain other components. In addition to all four deoxynucleotide triphosphates (dATP, dTTP, dCTP, dGTP), a buffer component and salts are added to it to set the appropriate ionic strength of the environment. An essential component is magnesium ions, which act as a polymerase cofactor. A too low concentration of Mg 2+ can result in a low yield of the reaction, on the contrary, a too high concentration can lead to the formation of non-specific products. Other components of the reaction mixture (albumin, ammonium sulfate, betaine, etc.) can stabilize the polymerase or improve the specificity of the product.
Some PCR modifications[edit | edit source]
Hot start[edit | edit source]
DNA polymerase is active even at temperatures lower than its temperature optimum, although the rate of the catalyzed reaction is considerably lower. After the reaction mixture is mixed at low temperature, the primers can hybridize to non-specific sites of the template and the polymerase can synthesize non-specific products from them until PCR is initiated and denaturation occurs. The formation of non-specific products can be prevented by not adding any of the reaction components to the reaction mixture and adding them only after heating to the denaturation temperature. So that the test tubes do not have to be opened, this separated component can be placed, for example, in a wax ball, which dissolves after heating. Another, the most widely used option today, is the use of antibody-inactivated DNA polymerase. After increasing the temperature, the antibody is denatured and thus released from the enzyme.
"Landing" (touchdown) PCR[edit | edit source]
If a large number of non-specific products are formed during PCR, their formation can be limited by using touchdown PCR. In the first cycles, a higher hybridization temperature is used than would correspond to the selected primers. The primers will be more difficult to bind to the template, i.e. the yield of the reaction will be lower, but they will bind very precisely and only a specific product will be formed. In subsequent cycles, the hybridization temperature is gradually reduced. The excess of the specific product over the original template already ensures the specificity of the reaction, and due to the better annealing of the primers at a lower temperature, a sufficient amount of product is formed.
Amplification of long chains[edit | edit source]
The most common polymerase used for PCR, Taq polymerase, cannot synthesize chains longer than about 2000 base pairs; after a certain time it is released from the template DNA and synthesis is interrupted. In addition, it cannot repair a misplaced nucleotide, so that a significant number of errors would occur in the synthesis of long chains. If it is necessary to amplify a long section of DNA (up to 30-40 thousand bases), a mixture of several thermostable DNA polymerases is usually used, and as a rule, enzymes with and without repair activity are combined. In the Anglophone literature, PCR of long chains is referred to as long distance PCR (LD-PCR).
Reamplification[edit | edit source]
If only a very small amount of template DNA is available, the yield of conventional PCR may not be sufficient. In that case, part of the PCR product can be used as a template for another PCR - we are talking about reamplification. Amplification of DNA in the first PCR can sometimes only be achieved at the cost of non-specific products. In order to amplify only the desired, specific fragments in the second PCR, a different pair of primers is often used, which lies between the primers used for the first amplification (i.e, in the second PCR a shorter section is amplified than in the first). This approach is referred to as nested PCR.
Allele-specific PCR[edit | edit source]
PCR itself can be used to detect a certain allele or to search for a mutation. The condition is that a primer must be available for the section in which the given allele differs from the others, or in which the sought mutation lies. If the examined DNA contains the corresponding sequence, amplification will proceed normally and a detectable amount of product will be produced at the end of the PCR. Otherwise, the primer will not attach and the DNA segment will not be amplified. The reaction must of course be supplemented with appropriate positive and negative controls.
Preparation of single-stranded DNA[edit | edit source]
For some techniques, it is necessary to obtain single-stranded DNA. This can be achieved by so-called asymmetric PCR, in which different amounts of primers are inserted into the reaction (usually in a ratio of 50:1 to 100:1). During PCR, double-stranded DNA will first be created in the usual way, which will increase exponentially. After about 20 cycles, however, the "minority," limiting primer is depleted. In subsequent cycles, only one strand will be formed (starting with the "excess" primer) and this single strand product will increase linearly.
Another option is to perform PCR with one primer labeled with biotin. The PCR product is then denatured and purified on a solid phase (e.g. agarose column) to which streptavidin is bound. Biotin has a high affinity for streptavidin, so the labeled strand is captured on the solid phase, while the other strand and the rest of the reaction mixture wash away.
PCR coupled to reverse transcription (RT-PCR)[edit | edit source]
RT-PCR is used when mRNA needs to be amplified instead of DNA . The first step is to isolate total RNA or mRNA from a tissue sample. This is followed by conversion of mRNA to cDNA using reverse transcriptase and then conventional PCR. Although the actual transcription of RNA into cDNA is not particularly difficult, this method is technically significantly more demanding than DNA amplification itself: RNA is rapidly degraded by ribonucleases, which are general contaminants of samples, tools and chemicals. In addition, RNases are thermostable, they are not damaged even by sterilization by autoclaving, and they retain their activity even after cleaning the material with some denaturing agents. To prevent RNA degradation, tools and solutions are treated with RNase inhibitors (e.g. diethyl pyrocarbonate, DEPC).
Quantitative PCR[edit | edit source]
In its basic configuration, the polymerase chain reaction is mainly used for qualitative determinations. The fact that after a certain number of cycles the rate of amplification slows down (see above) makes it difficult to use for the quantification of a certain sequence in a sample (eg to measure the expression of a gene). Nevertheless, there are procedures that allow template quantification. They are usually based on measuring the amount of PCR product during amplification (so-called real-time PCR). Since sampling the reaction mixture would easily lead to contamination and inaccuracies, procedures are used to determine the synthesis of the product without opening the tube.
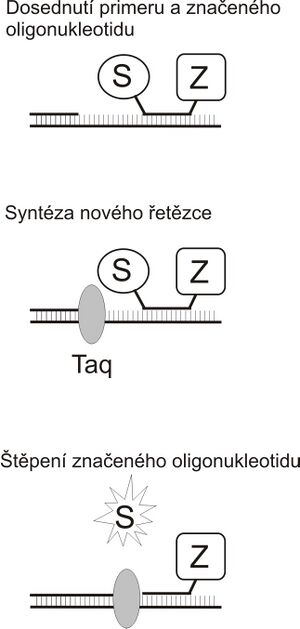
TaqMan PCR[edit | edit source]
An example can be the so-called TaqMan PCR. In addition to the primers, another oligonucleotide is inserted into the reaction, which adjoins the amplified section. This oligonucleotide is marked with a fluorescent marker at one end (marked S in the figure) and carries a so-called quencher (Z) at the other end. If the fluorescent substance is in close proximity to the quencher, its fluorescence is suppressed. The PCR itself proceeds in the usual way until the moment when the DNA polymerase encounters a labeled nucleotide during the synthesis of a new chain. At that point, it begins to displace it from the template strand and cleave it (Taq polymerase does not have a repair 3'-exonuclease activity, but it does have a 5'-exonuclease activity that allows degradation of the "interfering" strand). This releases the fluorescent probe into the solution and it is possible to measure the fluorescence directly in the tube during amplification. Fluorescence intensity is proportional to the amount of synthesized PCR product.
Multiplex polymerase chain reaction[edit | edit source]
Multiplex polymerase chain reaction (Multiplex PCR) is used to simultaneously amplify several different DNA sequences (as if many separate PCR reactions were performed, all together in one reaction). This process amplifies the DNA in the samples using several different primer pairs. Their design must be optimized so that all primer pairs can work at the same annealing temperature during PCR.
Multiplex PCR was first described in 1988. In 2020, multiplex RT-PCR assays were designed to combine multiple gene targets from the Centers for Disease Control in a single reaction to increase the availability and throughput of molecular testing for the diagnosis of SARS-CoV-2 .
Multiplex-PCR consists of multiple sets of primers within a single PCR mix to produce amplicons of different sizes that are specific for different DNA sequences. By targeting multiple sequences at once, more information can be obtained from a single test that would otherwise require multiple reagents and more time to perform. The annealing temperatures for each set of primers must be optimized to perform correctly within a single reaction, and the sizes of the amplicons, i.e. their base pair lengths, should be sufficiently different to produce distinct bands when visualized by gel electrophoresis. For use in real-time PCR, different amplicons are differentiated and visualized using primers that have been stained with fluorescent dyes of different colors. Commercial multiplex PCR kits are available and used by many forensic laboratories to amplify degraded DNA samples.
obrázek: https://www.nature.com/articles/s41598-018-37732-y/figures/2
Links[edit | edit source]
External links[edit | edit source]
- Wikipedia:PCR (english)
- Wikipedia:cs:Polymerase chain reactions
- https://en.wikipedia.org/wiki/Multiplex_polymerase_chain_reaction
Reference[edit | edit source]
- ↑ MULLIS, K B. The unusual origin of the polymerase chain reaction. Sci Am [online]. 1990, vol. 262, no. 4, p. 56-61, 64-5, Available from <http://www.ciens.ucv.ve:8080/generador/sites/labgeneticageneral/archivos/seminario%208-%20The%20Unusual%20Origin%20of%20PCR.pdf>. ISSN 0036-8733.
Resources[edit | edit source]
- ŠMARDA, Jan. Metody molekulární biologie. 1. edition. Masarykova univerzita, 2005. ISBN 80-210-3841-1.