Degradative system of the cell
Lysosomes[edit | edit source]
Site of intracellular digestion and exchange of cellular components. Vesicles endowed with a membrane containing the entire spectrum of most often hydrolytic enzymes come from the RER (the fact that they belong to lysosomes can be recognized by the bound signal molecule on their vesicle - mannose-6-phosphate). They are found in large quantities in phagocytic cells. Basically, they are active in acidic pH.
- PRIMARY (0,05–0,5 μm) – primary lysosomes are those that have not yet entered the digestion process (difficult to demonstrate in the cell);
- Secondary (0,2–2 μm);
- phagosome = primary lysosome + engulfed material;
- phagolysosome = secondary lysosome;
- autophagosome – lysosomes also replace worn-out organelles, after the fusion of the organelle with the primary lysosome, an autophagosome is formed;
- heterophagy – material is absorbed into the autophagic vacuole, it enters the cell from the environment. Lysosomes then fuse with this vacuole and empty their hydrolytic enzymes into the vacuole → digestion follows and this composite structure is referred to as a secondary lysosome;
- residual bodies = undigested residues remaining inside vacuoles;
- lipofuchsin = pigment from wear and tear, is stored in some long-lived cells as deposits of residual bodies.
Peroxisomes[edit | edit source]
Round membrane-bounded organelles 0,2–1 μm in size. They are formed in the endoplasmic reticulum. They contain enzymes for the degradation of hydrogen peroxide - for example [[ catalase]] (an enzyme that breaks down hydrogen peroxide into water and oxygen). They are the site of fatty acid degradation. They reduce O2 and H2O2. They protect the cell from the effect of hydrogen peroxide - which can cause damage to the cell.
Cytoplasmic inclusions[edit | edit source]
Usually transient components of the cytoplasm, formed by accumulated metabolites or deposits of various origins. They exist in the cell in several forms: fat droplets, glycogen (clusters of carbohydrates).
- Secretory granules – a form of proteins stored in gland cells, are periodically released into the extracellular media;
- pigments – deposits of colored substances, either synthesized inside the cell - melanin or coming from the outside carotene
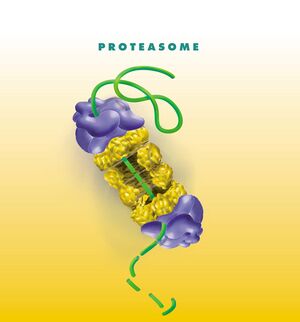
Proteasome[edit | edit source]
Characteristics[edit | edit source]
A protein that carries a K48 polyubiquitin chain. It can be recognized by the so-called 26S proteasome and degraded in it.
The 26S proteasome is a common type of proteasome found in our cells. It consists of two basic parts [1]:
- 20S proteasome, i.e. the core particle, which has the shape of a cylinder and in which the proteolysis of PDG itself takes place;
- 19S proteasome or regulatory particle, which is also called PA700.
Main particles[edit | edit source]
The 20S proteasome consists of a total of four rings. The two outer ones are formed by seven α units, and the two inner ones by seven β subunits. The active protein-cleaving sites are in the β rings and face the inside of the 20S proteasome cylinder, namely:
- β1 subunit with caspase-like activity ;
- β2 subunit with trypsin-like activity ;
- β5 subunit with chymotrypsin-like activity.
In addition to the normal 20S proteasome, inducible proteasomes also exist in our cells. These have other active sites (β1i, β2i and β5i) called immunoproteasomes or mixed proteasomes. They play a role in the immune response of cells to foreign substances[2]. A very special type of proteasome exists in the thymus, the so-called thymoproteasome . They contain a β5t subunit with unusual catalytic activity. Their role is related to the positive selection of CD8+ T cells. [3]
Regulatory particles[edit | edit source]
The proteasome Regulatory particles bind to the outer, i.e. α, ring of the 20S proteasome. In addition to the 19S proteasome, these can also be other complexes, such as PA28 or PA200, or even proteins that reversibly attach to the 20S proteasome in substoichiometric amounts [4]. Although the organization of proteasomes varies dynamically, it has been shown that the 26S proteasome remains intact during protein degradation [5] .
The regulatory particle of the 26S proteasome (PA700) contains two basic, interconnected regions: the base and the lid . In the base we can find six different AAA+ ATPases and another four subunits. Its main mission is to regulate entry into the interior of the 20S proteasome [6] . The lid contains nine non-ATPase subunits [7] and its basic function is the deubiquitination of ubiquitinated proteins by the JAMM domain DUB Poh1 before their entry into the interior of 26S proteasomes [8] .
Degradation of non-ubiquitinated proteins[edit | edit source]
A typical protein, degraded in a eukaryotic cell by the proteasome, must be ubiquitinated. However, according to recent findings, about 20% of all proteins cleaved by proteasomes in eukaryotic cells may not have ubiquitin labeling. Such proteins contain poorly ordered sites in their structures, which serve as a non-specific signal for degradation in proteasomes without the need for ubiquitination of the given protein.[9]
Degradation of ubiquitinated proteins[edit | edit source]
We will focus on the mechanism of degradation of ubiquitinated PDGs in 26S proteasomes.
Some subunits from the base (ubiquitin receptors) and some proteins that only transiently associate with 26S proteasomes play a key role in recognizing the ubiquitinated protein[10]. If the ubiquitinated protein is already bound to the 26S proteasome, its polyubiquitin chain can be variously cleaved by the proteasome and resynthesized by deubiquitinases and ubiquitin ligases [11] . It has also been shown that the reduction in the intensity of the degradation of ubiquitins themselves in the proteasome is related to the activity of a specific DUB, called Ubp6, which is not a constant subunit of the 26S proteasome [12] .
Single steps[edit | edit source]
- Before PDG degradation, the polyubiquitin chain is cleaved en bloc (as a whole, at once) by Poh1 and further removed by other DUBs [13].
- The unfolding of the protein into the primary structure and its movement into the opening of the proteasome is then associated with the hydrolysis of ATP by AAA+ ATPases.[14]
- The unfolded protein can be "stored" in the α rings if the β rings are still occupied by the degradation of the previous PDG.[15] Proteins can enter the 20S proteasome from both sides.[16] The degradation continues as long as the resulting oligopeptides are not small enough to diffuse out spontaneously.[17]
- As soon as the oligopeptides get out of the 26S proteasomes, they are further cleaved in the cell by other peptidases to amino acids that can be used for further proteosynthesis [18] , or are used within the immune system as antigens.[19]
Regulation of protein activity[edit | edit source]
Some proteins are not completely degraded by 26S proteasomes but are actually activated. This happens by degrading other proteins that are bound to them and inhibiting them. A typical example is an activation of the so-called nuclear factor-κB (NF-κB), which normally occurs in the cytoplasm in a complex with its inhibitor I-κB. Once this I-κB is ubiquitinated and degraded, NF-κB translocates to the nucleus and triggers the transcription of the relevant genes.[20] The function of 26S proteasomes is not only connected with the regulation of the amount of a given protein in the cell, but also with the regulation of the activity of various proteins. This implies that the UPS plays a key role in many therapeutically relevant processes, such as inflammatory diseases, neurodegenerative processes, muscular dystrophies, viral infections, or carcinogenesis -.[21][22]
Links[edit | edit source]
References[edit | edit source]
- BEDFORD, Lynn, Simon PAINE and Paul W SHEPPARD, et al. Assembly, structure, and function of the 26S proteasome. Trends Cell Biol [online] . 2010, vol. 20, no. 7, pp. 391-401, also available from < https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2902798/?tool=pubmed >. ISSN 0962-8924 (print), 1879-3088.
- ↑ STREHL, Britta, Ulrike SEIFERT, and Elke KRÜGER, et al. Interferon-gamma, the functional plasticity of the ubiquitin-proteasome system, and MHC class I antigen processing. Immunol Rev [online] . 2005, vol. 207, pp. 19-30, also available from < https://www.ncbi.nlm.nih.gov/pubmed/16181324 >. ISSN 0105-2896.
- ↑ MURATA, Shigeo, Yousuke TAKAHAMA, and Keiji TANAKA. Thymoproteasome: probable role in generating positively selecting peptides. Curr Opin Immunol [online] . 2008, vol. 20, no. 2, pp. 192-6, also available from < https://www.ncbi.nlm.nih.gov/pubmed/18403190 >. ISSN 0952-7915.
- ↑ DEMARTINO, George N and Thomas G GILLETTE. Proteasomes: machines for all reasons. Cell [online] . 2007, vol. 129, no. 4, pp. 659-62, also available from < https://www.ncbi.nlm.nih.gov/pubmed/17512401 >. ISSN 0092-8674.
- ↑ KRIEGENBURG, Franziska, Michael SEEGER, and Yasushi SAEKI, et al. Mammalian 26S proteasomes remain intact during protein degradation. Cell [online] . 2008, vol. 135, no. 2, pp. 355-65, also available from < https://www.ncbi.nlm.nih.gov/pubmed/18957208 >. ISSN 0092-8674 (print), 1097-4172.
- ↑ LI, Xiaohua and George N DEMARTINO. Variably modulated gating of the 26S proteasome by ATP and polyubiquitin. Biochem J [online] . 2009, vol. 421, no. 3, pp. 397-404, also available from < https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2872633/?tool=pubmed >. ISSN 0264-6021 (print), 1470-8728.
- ↑ SHARON, Michal, Thomas TAVERNER, and Xavier I AMBROGGIO, et al. Structural organization of the 19S proteasome lid: insights from MS of intact complexes. PLoS Biol [online] . 2006, vol. 4, no. 8, p. e267, also available from < https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1523230/?tool=pubmed >. ISSN 1544-9173 (print), 1545-7885.
- ↑ VERMA, Rati, L ARAVIND, and Robert OANIA, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science [online] . 2002, vol. 298, no. 5593, pp. 611-5, also available from < https://www.ncbi.nlm.nih.gov/pubmed/12183636 >. ISSN 0036-8075 (print), 1095-9203.
- ↑ BAUGH, James M, Ekaterina G VIKTOROVA, and Evgeny V PILIPENKO. Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. J Mol Biol [online] . 2009, vol. 386, no. 3, pp. 814-27, also available from < https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2649715/?tool=pubmed >. ISSN 0022-2836 (print), 1089-8638.
- ↑ VERMA, Rati, Robert OANIA, and Johannes GRAUMANN, et al. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell [online] . 2004, vol. 118, no. 1, pp. 99-110, also available from < https://www.ncbi.nlm.nih.gov/pubmed/15242647 >. ISSN 0092-8674.
- ↑ CROSAS, Bernat, John HANNA, and Donald S. KIRKPATRICK, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell [online] . 2006, vol. 127, no. 7, pp. 1401-13, also available from < https://www.ncbi.nlm.nih.gov/pubmed/17190603 >. ISSN 0092-8674.
- ↑ HANNA, John, Alice MEIDES, and Dan Phoebe ZHANG, et al. A ubiquitin stress response induces altered proteasome composition. Cell [online] . 2007, vol. 129, no. 4, pp. 747-59, also available from < https://www.ncbi.nlm.nih.gov/pubmed/17512408 >. ISSN 0092-8674.
- ↑ KOULICH, Elena, Xiaohua LI, and George N DEMARTINO. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol Biol Cell [online] . 2008, vol. 19, no. 3, pp. 1072-82, also available from < https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2262970/?tool=pubmed >. ISSN 1059-1524 (print), 1939-4586.
- ↑ STRIEBEL, Frank, Wolfgang KRESS, and Eilika WEBER-BAN. Controlled destruction: AAA+ ATPases in protein degradation from bacteria to eukaryotes. Curr Opin Struct Biol [online] . 2009, vol. 19, no. 2, pp. 209-17, also available from < https://www.ncbi.nlm.nih.gov/pubmed/19362814 >. ISSN 0959-440X (print), 1879-033X.
- ↑ SHARON, Michal, Susanne WITT, and Karin FELDERER, et al. 20S proteasomes have the potential to keep substrates in store for continuous degradation. J Biol Chem [online] . 2006, vol. 281, no. 14, pp. 9569-75, also available from < https://www.ncbi.nlm.nih.gov/pubmed/16446364 >. ISSN 0021-9258.
- ↑ HUTSCHENREITER, Silke, Ali TINAZLI, and Kirstin MODEL, et al. Two-substrate association with the 20S proteasome at the single-molecule level. EMBO J [online] . 2004, vol. 23, no. 13, pp. 2488-97, also available from < https://www.ncbi.nlm.nih.gov/pmc/articles/PMC449772/?tool=pubmed >. ISSN 0261-4189.
- ↑ KÖHLER, A, P CASCIO and DS LEGGETT, et al. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol Cell [online] . 2001, vol. 7, no. 6, pp. 1143-52, also available from < https://www.ncbi.nlm.nih.gov/pubmed/11430818 >. ISSN 1097-2765.
- ↑ VABULAS, Ramunas M and F Ulrich HARTL. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science [online] . 2005, vol. 310, no. 5756, pp. 1960-3, also available from < https://www.ncbi.nlm.nih.gov/pubmed/16373576 >. ISSN 0036-8075 (print), 1095-9203.
- ↑ KLOETZEL, Peter M. Generation of major histocompatibility complex class I antigens: functional interplay between proteasomes and TPPII. Nat Immunol [online] . 2004, vol. 5, no. 7, pp. 661-9, also available from < https://www.ncbi.nlm.nih.gov/pubmed/15224091 >. ISSN 1529-2908.
- ↑ RAPE, Michael and Stefan JENTSCH. Productive RUPture: activation of transcription factors by proteasomal processing. Biochim Biophys Acta [online] . 2004, vol. 1695, no. 1-3, pp. 209-13, also available from < https://www.ncbi.nlm.nih.gov/pubmed/15571816 >. ISSN 0006-3002.
- ↑ RUBINSZTEIN, David C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature [online] . 2006, vol. 443, no. 7113, pp. 780-6, also available from < https://www.ncbi.nlm.nih.gov/pubmed/17051204 >. ISSN 0028-0836 (print), 1476-4687.
- ↑ SCHWARTZ, Alan L, and Aaron CIECHANOVER. Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu Rev Pharmacol Toxicol [online] . 2009, vol. 49, pp. 73-96, also available from < https://www.ncbi.nlm.nih.gov/pubmed/18834306 >. ISSN 0362-1642.
- ↑ BEDFORD, Lynn, Simon PAINE and Paul W SHEPPARD, et al. Assembly, structure, and function of the 26S proteasome. Trends Cell Biol [online] . 2010, vol. 20, no. 7, pp. 391-401, also available from < https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2902798/?tool=pubmed >. ISSN 0962-8924 (print), 1879-3088.
- ↑ ↑ STREHL, Britta, Ulrike SEIFERT, and Elke KRÜGER, et al. Interferon-gamma, the functional plasticity of the ubiquitin-proteasome system, and MHC class I antigen processing. Immunol Rev [online] . 2005, vol. 207, pp. 19-30, also available from < https://www.ncbi.nlm.nih.gov/pubmed/16181324 >. ISSN 0105-2896.
- ↑ ↑ MURATA, Shigeo, Yousuke TAKAHAMA, and Keiji TANAKA. Thymoproteasome: probable role in generating positively selecting peptides. Curr Opin Immunol [online] . 2008, vol. 20, no. 2, pp. 192-6, also available from < https://www.ncbi.nlm.nih.gov/pubmed/18403190 >. ISSN 0952-7915.
- ↑ ↑ DEMARTINO, George N and Thomas G GILLETTE. Proteasomes: machines for all reasons. Cell [online] . 2007, vol. 129, no. 4, pp. 659-62, also available from < https://www.ncbi.nlm.nih.gov/pubmed/17512401 >. ISSN 0092-8674.
- ↑ ↑ KRIEGENBURG, Franziska, Michael SEEGER, and Yasushi SAEKI, et al. Mammalian 26S proteasomes remain intact during protein degradation. Cell [online] . 2008, vol. 135, no. 2, pp. 355-65, also available from < https://www.ncbi.nlm.nih.gov/pubmed/18957208 >. ISSN 0092-8674 (print), 1097-4172.
- ↑ ↑ LI, Xiaohua and George N DEMARTINO. Variably modulated gating of the 26S proteasome by ATP and polyubiquitin. Biochem J [online] . 2009, vol. 421, no. 3, pp. 397-404, also available from < https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2872633/?tool=pubmed >. ISSN 0264-6021 (print), 1470-8728.
- ↑ ↑ SHARON, Michal, Thomas TAVERNER, and Xavier I AMBROGGIO, et al. Structural organization of the 19S proteasome lid: insights from MS of intact complexes. PLoS Biol [online] . 2006, vol. 4, no. 8, p. e267, also available from < https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1523230/?tool=pubmed >. ISSN 1544-9173 (print), 1545-7885.
- ↑ ↑ VERMA, Rati, L ARAVIND, and Robert OANIA, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science [online] . 2002, vol. 298, no. 5593, pp. 611-5, also available from < https://www.ncbi.nlm.nih.gov/pubmed/12183636 >. ISSN 0036-8075 (print), 1095-9203.
- ↑ ↑ BAUGH, James M, Ekaterina G VIKTOROVA, and Evgeny V PILIPENKO. Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. J Mol Biol [online] . 2009, vol. 386, no. 3, pp. 814-27, also available from < https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2649715/?tool=pubmed >. ISSN 0022-2836 (print), 1089-8638.
- ↑ ↑ VERMA, Rati, Robert OANIA, and Johannes GRAUMANN, et al. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell [online] . 2004, vol. 118, no. 1, pp. 99-110, also available from < https://www.ncbi.nlm.nih.gov/pubmed/15242647 >. ISSN 0092-8674.
- ↑ ↑ CROSAS, Bernat, John HANNA, and Donald S. KIRKPATRICK, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell [online] . 2006, vol. 127, no. 7, pp. 1401-13, also available from < https://www.ncbi.nlm.nih.gov/pubmed/17190603 >. ISSN 0092-8674.
- ↑ ↑ HANNA, John, Alice MEIDES, and Dan Phoebe ZHANG, et al. A ubiquitin stress response induces altered proteasome composition. Cell [online] . 2007, vol. 129, no. 4, pp. 747-59, also available from < https://www.ncbi.nlm.nih.gov/pubmed/17512408 >. ISSN 0092-8674.
- ↑ ↑ KOULICH, Elena, Xiaohua LI, and George N DEMARTINO. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol Biol Cell [online] . 2008, vol. 19, no. 3, pp. 1072-82, also available from < https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2262970/?tool=pubmed >. ISSN 1059-1524 (print), 1939-4586.
- ↑ ↑ STRIEBEL, Frank, Wolfgang KRESS, and Eilika WEBER-BAN. Controlled destruction: AAA+ ATPases in protein degradation from bacteria to eukaryotes. Curr Opin Struct Biol [online] . 2009, vol. 19, no. 2, pp. 209-17, also available from < https://www.ncbi.nlm.nih.gov/pubmed/19362814 >. ISSN 0959-440X (print), 1879-033X.
- ↑ ↑ SHARON, Michal, Susanne WITT, and Karin FELDERER, et al. 20S proteasomes have the potential to keep substrates in store for continuous degradation. J Biol Chem [online] . 2006, vol. 281, no. 14, pp. 9569-75, also available from < https://www.ncbi.nlm.nih.gov/pubmed/16446364 >. ISSN 0021-9258.
- ↑ ↑ HUTSCHENREITER, Silke, Ali TINAZLI, and Kirstin MODEL, et al. Two-substrate association with the 20S proteasome at the single-molecule level. EMBO J [online] . 2004, vol. 23, no. 13, pp. 2488-97, also available from < https://www.ncbi.nlm.nih.gov/pmc/articles/PMC449772/?tool=pubmed >. ISSN 0261-4189.
- ↑ ↑ KÖHLER, A, P CASCIO and DS LEGGETT, et al. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol Cell [online] . 2001, vol. 7, no. 6, pp. 1143-52, also available from <
- ↑ ↑ VABULAS, Ramunas M and F Ulrich HARTL. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science [online] . 2005, vol. 310, no. 5756, pp. 1960-3, also available from < https://www.ncbi.nlm.nih.gov/pubmed/16373576 >. ISSN 0036-8075 (print), 1095-9203.
- ↑ ↑ KLOETZEL, Peter M. Generation of major histocompatibility complex class I antigens: functional interplay between proteasomes and TPPII. Nat Immunol [online] . 2004, vol. 5, no. 7, pp. 661-9, also available from < https://www.ncbi.nlm.nih.gov/pubmed/15224091 >. ISSN 1529-2908.
- ↑ ↑ RAPE, Michael and Stefan JENTSCH. Productive RUPture: activation of transcription factors by proteasomal processing. Biochim Biophys Acta [online] . 2004, vol. 1695, no. 1-3, pp. 209-13, also available from < https://www.ncbi.nlm.nih.gov/pubmed/15571816 >. ISSN 0006-3002.
- ↑ ↑ RUBINSZTEIN, David C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature [online] . 2006, vol. 443, no. 7113, pp. 780-6, also available from < https://www.ncbi.nlm.nih.gov/pubmed/17051204 >. ISSN 0028-0836 (print), 1476-4687.
- ↑ ↑ SCHWARTZ, Alan L, and Aaron CIECHANOVER. Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu Rev Pharmacol Toxicol [online] . 2009, vol. 49, pp. 73-96, also available from < https://www.ncbi.nlm.nih.gov/pubmed/18834306 >. ISSN 0362-1642