Searching for mutations
Mutation search techniques are methods that make it possible to find individuals with rare polymorphisms or previously unknown mutations in the population . They can also be used to determine whether certain stretches of DNA obtained from different individuals are the same or different from each other. When searching for mutations, the procedure is actually similar: a section of DNA from many different individuals is compared with reference DNA (wild allele of a gene, etc.).
Conformation polymorphism of single chains[edit | edit source]
This is a technique for searching for mutations in the genome. It is among the simplest techniques for this activity. It is often denoted by the abbreviation SSCP (single strand conformation polymorphism) .
Principle of technique[edit | edit source]
Its principle is electrophoresis of single-stranded DNA on a non-denaturing polyacrylamide gel at low temperature. The DNA strand "packs" according to internal complementarities, creating a spatial structure similar to e.g. tRNA. The speed at which single-stranded DNA then travels during nucleic acid electrophoresis depends on the exact conformation. Since even a very small change in the nucleotide sequence can cause the DNA to form a completely different spatial structure, it is often possible to differentiate even a single base substitution using SSCP .
Working with PCR[edit | edit source]
When working with most PCR products, the DNA is separated into two groups of fractions during SSCP: The first group travels more slowly, the bands tend to be sharper and there are more of them. The second faction travels quickly and usually forms a single lane.
There are several reasons why one PCR product forms several bands:
- each strand of denatured DNA assumes a different conformation,
- one chain can form several stable conformations,
- some of the molecules do not form a packed spatial structure,
- part of the molecules renatures into the original double helix and thus homoduplexes are formed again,
- in heterozygotes, part of the molecules can renature to form heteroduplexes.
Reliability of technology[edit | edit source]
It has been reported that 99% of point mutations can be captured by SSCP when working with a 100-300 bp stretch of DNA, over 80% for stretches of 400 bp. As the length of the examined DNA fragment increases, the efficiency of SSCP decreases, and this technique is not suitable for sections longer than about 750 bp.
DGGE[edit | edit source]
A very sensitive technique for searching for mutations is electrophoresis in a gradient denaturing gel (denaturing gradient gel electrophoresis, DGGE ). It uses the fact that the rate of DNA denaturation depends on the number of hydrogen bonds. DNA strands will separate more easily at sites rich in AT pairs, while stretches rich in CGs will be more stable. A polyacrylamide gel with a gradually increasing concentration of denaturing substances (formamide and urea) is used for electrophoresis. The examined DNA travels in the electric field at a speed corresponding to its molecular weight, until the moment when the two strands begin to separate from each other. The resulting denatured strands travel much more slowly during electrophoresis, so the easier a certain section is denatured, the closer to the start the DNA sample stops. Since completely separated ssDNA would create fuzzy bands, primers with a so-called CG-clamp are used for DNA amplification. The PCR product then contains double helices that have only CG pairs at one end; at this point the chains are not easily denatured. The sensitivity of DGGE is close to 100%.
TGGE[edit | edit source]
A similar technique is TGGE (temperature gradient gel electrophoresis ) . Instead of a gel with a gradually increasing concentration of denaturing substances, gradually increasing gel temperatures are used.
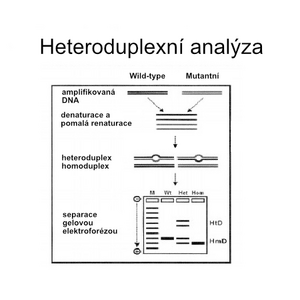
Heteroduplex analysis[edit | edit source]
Heteroduplex analysis is a method of direct molecular genetic diagnosis related to DGGE. The analyzed sample is mixed with reference DNA, denatured and then allowed to hybridize again.
If the sequence of the reference DNA differs from the sample (for example due to the presence of a mutation), in addition to the original homoduplexes, mixed molecules consisting of one strand from the sample and one from the reference DNA will be formed. Because such heteroduplexes contain a non-complementary stretch without hydrogen bonds, they will disintegrate significantly earlier during electrophoresis on a denaturing gel.
Another possibility is cleavage of heteroduplexes at the site of non-complementarity. This is possible chemically or, more reliably, enzymatically (eg T4 endonuclease VII or T7 endonuclease I). This approach is referred to as enzyme mismatch cleavage (EMC). It is reported to be 90% effective.
Currently, this type of analysis is practically no longer used and has been replaced by sequencing methods.