Control of gene expression and proteosynthesis in prokaryotes
The molecular basis of bacterial regulation of gene expression was recognized earlier and in more detail than eukaryotic regulation, because bacterial systems are simpler and because bacteria have a short cell cycle, which substantially speeds up genetic studies. In prokaryotes, regulation at the level of transcription predominates.
Regulation at the level of transcription[edit | edit source]
Regulation σ-factors[edit | edit source]
A promoter is a stretch of DNA at which RNA polymerase begins to synthesize a strand of RNA. One of the subunits of the bacterial RNA polymerase is the σ-factor, without it the enzyme can not recognize the promoter. Different types of σ-factors allow the polymerase to target different promoters. Factors are designated by molecular weight. Under optimal conditions, σ70 acts in the bacterium. During heat shock, other proteins are synthesized due to factor σ32. During phage infection, σ is activated in the bacterium, which helps start the transcription of so-called late genes. σ37 initiates transcription of sporulation genes. If the bacteria in the nutrient medium does not have enough nitrogenous substances, σ60 will ensure the use of other resources.
Jacob-Monod operon model[edit | edit source]
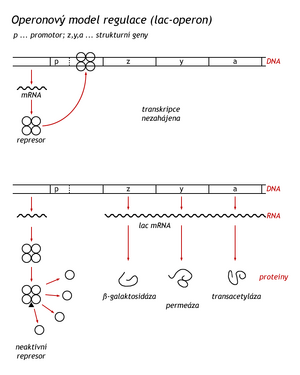
In 1961, Frenchmen Jacob and Monod published a theory explaining the mechanism by which bacteria change the spectrum of synthesized enzymes depending on the type of nutrients available to them. This theory of transcriptional regulation has been fully confirmed. The basic regulated unit on DNA is an operon. It consists of an operator and tandemly arranged structural genes. An operator is a DNA sequence to which a protein called a repressor binds.
To use lactose as an energy source, the bacterium needs the enzymes β-galactosidase, galactoside permease and β-galactoside transacetylase, which are encoded by the lactose operon (lac-operon). E. coli grown on soil with favorable energy sources, i.e. glucose or glycerol, does not need these enzymes. Under these circumstances, they are represented in the cell by only a few molecules. If E.coli is transferred to a nutrient medium, in which the mentioned energy sources will be replaced by lactose, the synthesis of enzymes required for its use is induced in the bacterium. Therefore, they are called inducible enzymes. The natural inducer is 1,6-allolactose, which is formed from lactose by transglycosylation, catalyzed by the few persistent β-galactosidase molecules.
When a constitutive mutant was discovered that synthesized all three enzymes regardless of available carbohydrates, Jacob and Monod predicted that the synthesis of all three enzymes was controlled by a single regulation gene which diffusible product (protein repressor) would bind to the operator and prevent the synthesis of the polygenic (polycistronic) mRNA. Today we know that ribosomes and translation factors bind to this mRNA during transcription and individual enzymes are synthesized, not the protein precursor corresponding to the entire operon. The synthesis of long protein precursors and their proteolytic cleavage into functional peptides is a phenomenon observed in eukaryotic cells.
Lac repressor (Mr=37000) consists of four identical subunits. The tetramer has a high affinity for the operator, to which it binds and stops transcription of the operon. After four inducer molecules are bound, the repressor dissociates into subunits and is released from the DNA. Synthesis of mRNA and enzymes can be initiated. Repressors are characterized by high affinity to operators. The dissociation constant of the lac-repressor-operator complex is 10 M. The repressor first binds to a non-specific DNA site and travels along the DNA. So it searches for the operator one-dimensionally, along the DNA. Binding the repressor three-dimensionally, directly from solution, would be tedious. The operator sequence (27 bp) is centrally symmetrical, which is common in stretches of DNA designed for specific protein binding.
Regulatory significance of cAMP in bacteria[edit | edit source]
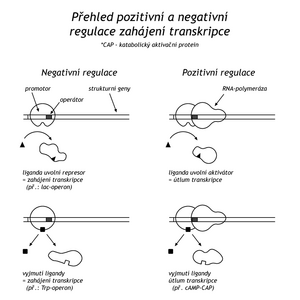
Regulation on the lactose operon is an example of negative regulation of transcription, meaning that genes are transcribed until transcription is stopped by a repressor protein. In positive regulation, genes are active only if the regulatory protein is bound to DNA. An example of positive regulation is catabolic repression.
If glucose is available to E.coli, the bacterium has a low level not only of β-galactosidase, but also of other catabolic enzymes (galactokinase, tryptophanase, arabinoisomerase). In the absence of glucose, the concentration of cAMP rises in the cell, which binds to the catabolically activating protein (CAP-protein, catabolite activating protein). The CAP-cAMP complex has a high affinity for the part of the promoters of individual genes of several catabolic enzymes. The complex binds in such a way that, unlike a repressor, it does not exceed the binding site for RNA polymerase, rather it creates additional binding sites and thus stimulates transcription.
Thus, two regulatory mechanisms operate at the transcription initiation site of the lactose operon. The repressor interferes with the initiation of transcription, CAP-cAMP facilitates it. If there is both glucose and lactose in the environment, the bacterium will prefer glucose, because the cAMP level in the cell will decrease due to glucose and the derepression of the lactose operon is not stimulated due to the lack of CAP-cAMP.
Variation in operon control of genes. Tryptophan and arabinose operon.[edit | edit source]
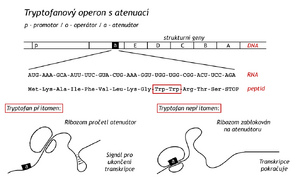
The tryptophan operon controls the synthesis of five enzymes required for the formation of tryptophan from chorismate. The repressor of the tryptophan operon, unlike the lactose repressor, can bind to the operator only in a complex with a co-repressor, which in this case is tryptophan. Only in this complex does the repressor adopt a conformation with high affinity for the operator. It is expedient, because if there is enough Trp in the environment, the bacterium has no reason to synthesize enzymes to create it. In the absence of tryptophan, the Trp-repressor-operator complex dissociates and thereby unblocks polycistronic mRNA synthesis.
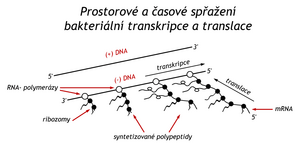
Spatial and temporal coupling of bacterial transcription and translation.
The lactose repressor controls the expression of a single operon, while the tryptophan repressor controls the function of three operons. It controls the aforementioned Trp-operon (5 enzymes), another operon for three enzymes of aromatic amino acid biosynthesis, and in addition regulates its own production on a regulatory gene. During the study of the Trp-operon, another level of transcription regulation, attenuation, was discovered. The regulatory signal in this case comes from the translational level of proteosynthesis. In a bacterium, this is made possible by the fact that transcription and translation are linked here both spatially and temporally.
The attenuator is the stretch of DNA between the operator and the gene for the first of a group of enzymes. It encodes the mRNA leader sequence at its 5'-end. This sequence is translated by the ribosome into a peptide whose 10th and 11th positions are tryptophans. The fourteenth codon of the leader sequence is the terminator codon. If there is enough Trp in the environment, the ribosomes "read" to the termination codon and the beginning of the mRNA conforms to a shape signaling the end of the synthesis of the translated (and not yet completed) mRNA. An mRNA hairpin is formed at the end of the attenuator, which is the terminator for RNA polymerase. For lack of Trp the ribosome stops at the codons for Trp and the 5'-end of the mRNA with the ribosome forms such a conformation that prevents the formation of a terminator for RNA polymerase. Transcription continues. It makes sense, bacteria need enzymes to synthesize Trp. Attenuation is known for a number of other operons (for Phe, His, etc. synthesis enzymes)
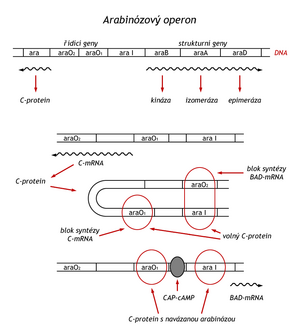
Some proteins can be both positive and negative regulators. Such a repressor controls, for example, the arabinose operon, which controls the synthesis of three enzymes that convert arabinose into xylulose-5-phosphate (araB, araA, araD). In this case, the regulatory gene C is adjacent to an operon, and it has two operators (ara O1 and ara O2) and one promoter (ara 1). As a catabolic operon, the arabinose operon is also controlled by the cAMP-CAP complex, facilitating the initiation of transcription. Three situations can occur:
- In the absence of regulatory protein C, mRNA for its synthesis is synthesized.
- Protein C synthesis is autoregulated. In excess of protein C in the absence of arabinose, protein C binds simultaneously to the O2 operator and another molecule of protein C to the O2 operator and the aral region, which is the promoter of structural genes. A DNA loop is formed and enzyme synthesis is blocked.
- If the level of cAMP in the cell rises when there is a lack of glucose and there is arabinose in the environment, arabinose binds to protein C. This changes its conformation into a positive regulatory factor, which, together with the cAMP-CAP complex, enables the start of mRNA synthesis for enzyme synthesis. The synthesis of protein C is still blocked by the arabinose-protein C complex bound to the O1 operator.
Even more complex regulatory systems are known, in which one gene after another is gradually activated programmatically in a certain sequence and after certain conditions have been reached. This is the case, for example, after infection of a cell with a virus (phage). In the first phase of infection, the host cell synthesizes factors and enzymes for the synthesis of viral components, and in the late phase of infection, the program changes in favor of the synthesis of its own virion components. Such a program was studied in detail, for example, with phage λ. The program is extremely efficient, because the virus is "allowed" to kill its host only after using its proteosynthetic mechanisms.
Control of transcription termination[edit | edit source]
During transcription, RNA polymerase moves along the DNA up to the terminator sequence. After its transcription, the synthesized RNA is twisted into a hairpin, which helps dissociate the RNA from the matrix and release the enzyme. After the hairpin there is a stretch of oligo (U) that binds to the matrix more weakly than the sequence containing G and C. There are proteins called anti-termination factors that allow the enzyme to "read" through the terminator sequence and continue to synthesize the longer RNA.
Termination of transcription could also be controlled by the level of ρ-factor (rho-factor). It is a protein, a certain concentration of which is needed to terminate transcription at certain terminators. These observations have not yet been confirmed in vivo .
Regulation of bacterial proteosynthesis at the level of translation[edit | edit source]
Translational repression is known in bacteria. An example is the synthesis of ribosomal proteins. The synthesis of fifty of these proteins is precisely coordinated on 20 operons. Polycistronic mRNA coding for a certain group of ribosomal proteins and sometimes other proteins (EF-G, EF-Tu, etc. RNA polymerases) is synthesized on each operon. One of the proteins in the group is always a repressor of translation. It binds to mRNA (not DNA!) and suppresses the synthesis of a whole group of proteins encoded on the mRNA molecule. This regulation is overridden by rRNA synthesis. The affinity of ribosomal proteins for rRNA is higher than for the binding site on mRNA. So as long as there is enough rRNA, more ribosomal proteins are synthesized. If the synthesis of rRNA is stopped or slowed down, an excess of ribosomal proteins - repressors begins to bind to the corresponding mRNA and stops translation (of itself and of other ribosomal proteins).
Links[edit | edit source]
Related arcticles[edit | edit source]
Recources[edit | edit source]
- ŠTÍPEK, Stanislav. Stručná biochemie : Uchování a exprese genetické informace. 1. edition. Medprint, 1998. 92 pp. pp. 54–62. ISBN 80-902036-2-0.