Genetic linkage
Definition[edit | edit source]
Genetic linkage is a phenomenon where certain alleles of different genes do not conform to the Mendel´s law of independent segregation because they are localized on the same chromosome, not very far from each other.
History[edit | edit source]
Linkage was discovered by Bateson and Punnett in 19XX in sweet pea (Lathyrus odoratus), but explained first by T. H. Morgan using fruitflies (see lecture el.lf1.cuni.cz/linkage).
Principle[edit | edit source]
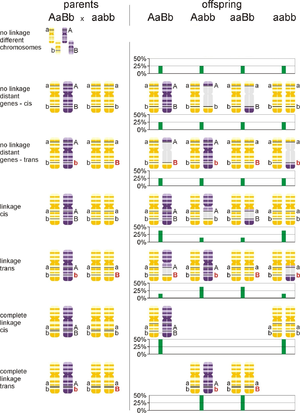
In backcross AaBb x aabb, if A and B genes segregate independently, The double heterozygote parent makes 4 gamete types AB, aB, Ab and ab, while the homozygous parent makes only ab gametes. Therefore offspring genotypes are AaBb, aaBb, Aabb and aabb. That makes 4 phenotypic groups in case of complete dominance, incomplete dominance and codominance, with equal probability 25% each, segregation ratio is 1:1:1:1. If A and B are localized on a single chromosome, and heterozygote parent AaBb is AB/ab (haplotypes “AB” and “ab”, cis-phase of linkage), then AaBb and aabb offspring groups will be more populated compared to Aabb and aaBb groups, which require a crossing over in AB/ab parent for their formation. These less common individuals are called recombinants. In trans-phase of linkage AaBb = Ab/aB and the situation is opposite compared to cis (AaBb and aabb are recombinants and they are less numerous). See Fig. 1).
Measuring gene distance[edit | edit source]
With increasing distance between the two genes, crossing-over is more likely to occur between the genes and the percentage of recombinants increases. recombination fraction = number of recombinants/number of all individuals percentage of recombinants = 100xrecombination fraction However, the percentage of recombinants does not increase linearly with physical distance, since it reaches a plateau of 50%. This is due to the fact, that as the distance is increasing, at the beginning probability increases for a single crossing over, but later also for a double crossing-over, which looks like no crossing-over, if only the two genes are sampled. Mathematical solution of the relationship between recombination fraction and distance leads to mapping function (Haldane, lecture, Kosambi etc.). However, the physical distance cannot be simply recalculated from the linkage distance, since there are many hotspots and coldspots on each chromosome with different crossing over probability, also that probability is higher for females and rises towards telomeres. A rule of thumb is 1 cM for 1 Mbp, variation is at least an order of magnitude.
Ordering loci[edit | edit source]
Linkage enables ordering of loci, if there are three or more. It is impossible to decide which locus is first and which is last (not surprisingly, in chromosomes this was set by convention), but between them the order can be solved. In the simplest version, a map is constructed from three loci in a “three point experiment”.
Genetic maps[edit | edit source]
By ordering loci that are linked, a genetic map is constructed, containing order and distance information. Reference linkage maps were constructed for human. Link to the human genome resources.
Polymorphic loci for genetic maps[edit | edit source]
can be divided into polymorphic phenotypes inherited in mendelian fashion and polymorphic DNA “markers”. Polymorphic phenotype can be the presence/absence of any mendelian disease, physiological traits like eye color, blood groups, HLA antigens, variants of serum proteins etc. Polymorphic DNA markers comprise single nucleotide polymorphisms (SNPs), tandem repeat polymorphisms and other insertion/deletion polymorphisms. Historically, many DNA polymorphisms are called by a predominant method of detection, e.g. subset of SNPs can be called “restriction fragment length polymorphisms”, since restriction endonucleases are used to assay the variable nucleotide.
Use of linkage[edit | edit source]
Indirect DNA diagnostics[edit | edit source]
is based on linkage of unknown mutation causing the disease and a known DNA polymorphism (rarely a polymorphic phenotype) that is in vicinity of the disease causal gene. A family is necessary with at least one affected (details differ based on mode of inheritance). Principle and examples see lecture, practical trainings.
Positional cloning[edit | edit source]
is a method for identification of genes causal for a certain genetic disease based on finding linkage between the disease phenotype and a certain chromosome region – where the causal mutations can then be searched. Without linkage, there are two other possibilities: for some genetic diseases, especially “metabolic disorders”, there can be a biochemical clue leading to identification of the responsible protein. Newest alternative is using the massively parallel sequencing methods to obtain sequence of complete genome or complete exome (all coding sequences) of the affected individual, and compare it to reference human genome.
Linkage disequilibrium[edit | edit source]
is a situation, where a haplotype is more represented in a population compared to expected value derived from the allele frequencies. This is usually because the loci are so close to each other, that crossovers occur very rarely, therefore the alleles of the haplotype are still more likely to occur together in the same arrangement as in a founder of that population. As a simplification, we can say that linkage is observed inheritance of a haplotype in a family (typically up to 4 generations), while linkage disequilibrium is the same phenomenon operating across multiple generations on whole population level. Not surprisingly, linkage in a family operates on a chromosome level, connecting genes up to tens of Mbp apart, while linkage disequilibrium operates on a much smaller scale measured in kbp.
Calculation of linkage disequilibrium[edit | edit source]
If haplotype AB has population frequency X, while allele A has frequency a and allele B frequency b, then linkage disequilibrium is D = x – a*b ≠ 0. The value D depends on allele frequency, so a “normalized” value is used D´ = D/Dmax, where Dmax = min(a,b).
