General structure of enzymes
Many different reactions take place in biological systems. Experiments have shown that the rate of these reactions when carried out outside a living system (i.e. ``in vitro) is much lower than ``in vivo. In living systems, reactions take place hundreds to millions of times faster than in vitro. It is caused by specific catalysts - enzymes'. These often enable the course of such reactions that would otherwise practically not take place in the conditions of the human body (temperature, pH, etc.).
Biological catalysts[edit | edit source]
A Catalyst' is a substance that increases the rate of a chemical reaction, but does not change the chemical equilibrium (only shortens the time it takes to achieve it). During the reactions, the enzyme molecule is not consumed.
Enzyme[edit | edit source]
An enzyme is a specific organic molecule that accelerates reactions in organisms (it acts as a biocatalyst). This enables reactions to take place even at relatively low temperatures, neutral pH and atmospheric pressure, which are normally found in organisms. The vast majority of enzymes are proteins. The exception is some types of RNA molecules - so-called ribozymes.
In addition to the protein component, enzymes can also contain a "non-protein component". According to its presence, enzymes can be divided into:
- simple':
- composite':
- in addition to the protein part (the so-called apoenzyme, it is ineffective by itself), they also contain a non-protein part - the so-called cofactor. The cofactor together with the apoenzyme forms an active enzyme molecule, the so-called 'holoenzyme.
Cofactors[edit | edit source]
A cofactor can be:
- metal ions: Zn2+ (e.g. alcohol dehydrogenase), Mn2+ (e.g. arginase), Fe2+, Cu2+, Mg2+,
- organic molecules: these are often derivatives of vitamins.
According to the nature of the binding to the apoenzyme, cofactors are divided into:
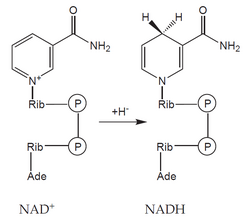
- coenzymes': an organic molecule of a non-protein nature, loosely bound to the apoenzyme molecule - it can be separated from it (e.g. NAD+, NADP+),
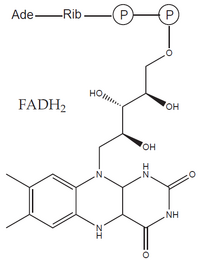
- prosthetic groups: an organic molecule of a non-protein nature, tightly bound to the apoenzyme molecule (eg heme, FAD).
Multienzyme Complex[edit | edit source]
An enzyme can be made up of a different number of peptide chains. Each chain can contain multiple domains (with the same or different enzyme specificity). If the enzyme contains several chains (quaternary structure), we refer to it as a multienzyme complex. Individual subunits usually have different specificities and are non-covalently linked to each other. An example of a multienzyme complex is fatty acid synthase, which catalyzes the synthesis of higher fatty acids in cells.
Zymogens[edit | edit source]
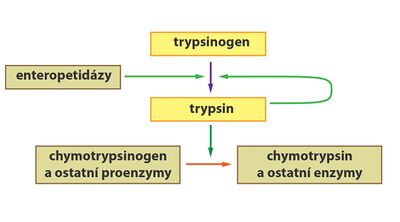
Some enzymes (eg digestive) are formed and secreted in their inactive form as so-called zymogens (proenzymes). The reason is the protection of synthesizing cells from being split by the effect of active forms of enzymes. Zymogens are activated only at the point where their activity is required. Activation can, for example, take place as so-called partial proteolysis, during which a precisely defined part of the proenzyme molecule is cleaved off.
Here are two examples of this process:
- The chief cells of the gastric mucosa secrete the proenzyme pepsinogen''. The HCl present in the gastric juice aids in the autoactivation of pepsinogen to active pepsin. The reaction also takes place autocatalytically, when already formed molecules pepsin participate in the splitting of pepsinogen.
- Like pepsin, trypsin is also synthesized in the pancreas as inactive trypsinogen. In the small intestine, the hexapeptide is subsequently cleaved with the help of the enzyme "enteropeptidase" (formed by the cells of the intestinal mucosa) to form active trypsin.