Gene Mutations
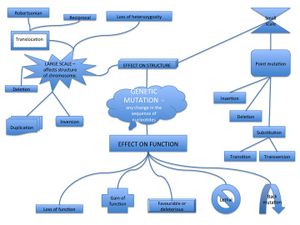
A mutation is a permanent heritable change in the nucleotide sequence in a gene or chromosome. A mutation can be caused by copying errors during DNA synthesis in the S phase of Interphase, by mutagens or they can be induced by the cell itself. Mutations can be catigorized according to their effect on the function of the gene product, or by their effect on the structure of the chromosome or gene.
Mutation by effect on the function of the gene product[edit | edit source]
- Loss of function mutation means that the function of the gene is lost and that it will not produce the same functional gene product, e.g. protein or enzyme. This mutation is often recessive and all genes that code for the same gene product need to be mutated for the mutation to have an effect on the phenotype.
- Gain of function mutation changes the gene product so that it will have a new or abnormal function. This mutation is usually dominant so only one mutation is enough for it to be phenotypically noticeable.
- Dominant negative mutation will act antagonistically (in opposition) to the normal gene function. This may have a severe effect on the phenotype since it will resemble a loss of function mutation. However the severity of the phenotypic effect may be reduced by increased translation of the "wild type" gene.
- Lethal mutation will simply lead to the death of the organism mainly by apoptosis.
- Back mutation will restore the original sequence of the gene to the "wild type" variant.
Another, more general way of categorizing a mutations effect is into favourable and deleterious mutations.
A favourable mutation would increase the fitness in mating of the carrier and this would eventually result in an evolutionary change.
Deleterious mutations cause the fitness of the carrier to be reduced and will eventually disappear or remain at a small frequency in the population through natural selection.
Effect on structure[edit | edit source]
The effect on structure can be divided into large scale mutations and small scale mutations.
Large scale mutations[edit | edit source]
Affect the structure of a whole chromosome or part of a chromosome. This type of mutation is often the effect of an error in S phase or during recombination in meiosis. The large scale mutations can be divided into:
- Duplication of genes.
- Deletions of one of the arms of the chromosome or a part of an arm. They can be either terminal which means that there is a deletion of one of the end regions of one arm, or they can be interstitial which is a deletion somewhere in the middle of one arm.
Translocations are caused by rearrangements of parts of non-homologous chromosomes. They can be either balanced or *unbalanced. A balanced translocation will not affect the carrier since no genetic information is lost. Translocations are also divided into Robertsonian translocation which is the fusion of two acrocentric chromosomes, and Reciprocal translocation which is due to an error during the Pachytene phase in Prophase I where structures called pachytene quadrivalent form and this results in errors in recombination.
- Inversions are when the physical orientation of the genes change and are flipped "up side down". They can be either paracentric (involve genes located in an arm of the chromosome, or they can be pericentric (genes are flipped around the centromere).
- Loss of heterozygosity is loss of one allele either by deletion or recombination. This is an important mechanism behind the "two-hit model" of cancer.
Small scale mutations[edit | edit source]
Affect only a single nucleotide and is usually the result of an mismatch error in S phase of Interphase or the result of DNA damage from UV-radiation from the sun. It is also called a point mutation. The consequences of point mutations can be silent, missense or nonsense mutations. Point mutations can be divided into:
- Substitution which is the exchange of a single nucleotide. If the exchange is a purine to a purine or pyrimidine to pyrimidine then it is called a transition. If the substituton is of a purine with a pyrimidine or opposite then it is termed transversion.
- Insertion/deletion is the adding or removal of one or more nucleotides. The result can be a frameshift mutation that changes the reading frame. Another effect might be a splice site mutation that occurs where splicing usually occurs. This would change the transcript so introns could be inserted to mature mRNA molecules.