Caspases
Caspases are a family of cysteine proteases with an active site that cleaves a peptide bond specifically behind aspartate. They were first discovered and studied on a model organism of the nematode (Caenorhabditis elegans, products of its ced - cell death abnormal genes ) in connection with ontogenesis during which apoptosis occurs physiologically (e.g. in synaptically isolated neurons, in some animals during finger development, etc.). For their importance in basic cellular functions, genes that encode them have maintained very conserved sequences throughout evolution.
Distribution[edit | edit source]
Twelve caspases are known in humans , seven of which occur in the apoptotic cascade, while the others are associated with cell cycle cytokines , inflammation , and other processes.
Caspases involved in the apoptotic cascade[edit | edit source]
- Initial –they are at the beginning of the activation cascade, they have a prodomain of 129–219 amino acid residues
- caspases -8 a -10:prior to activation, contain a sequence called the DED (death-effector domain ; about 80 AMK), which allows binding to an adapter protein that forms a complex of two caspases to allow mutual cleavage;
- caspase -2 a -9: contains CARD ( caspase recruitment domain ; approx. 90 AMK) sequence before activation
- The effectors – caspase -3, caspase -6 a caspase -7 – are the terminal part of the activating caspase, performing the activation of the apoptosis process itself.
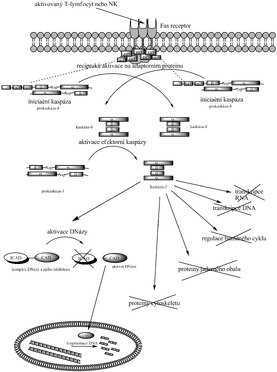
As they are used as a substrate by eg actins (thus changing the cytoskeleton), laminins (nuclear membrane), proteins related to cell cycle regulation (eg Wee1, cyclin A, ATM, p21 Cip , pRb), proteins involved in DNA replication (eg topoisomerase I, Mcm3), transcription (e.g. Sp-1, NF-κD) and signal transduction (e.g. RasGAP, kinase C). However, the morphological changes that cause these events have not yet been studied in detail. DNA degradation is also involved because they activate a DNase called CAD ( caspase activated DNase ) by cleaving ICAD ( inhibitor of caspase activated DNase).), which has been linked to CAD since protein folding and hinders its function. Upon release from the CAD-ICAD complex, CAD enters the nucleus and performs DNA fragmentation. Note: In addition to CARD and DED, sequences recognized by caspases include DD and others; they contain not only the caspases themselves, but also their adapter proteins, receptors, substrates, etc.
Activation mechanism[edit | edit source]
Caspases are expressed in an inactive form, as so-called procaspases - dimers, which in each measure have a part α and β arranged in sequence and an amino acid sequence attached before the molecule itself, called pro-domain (on one chain possibly replaced by 2 × DED or CARD) . Their specific feature is that one caspase can activate another. They are arranged in such a way that after cleavage of the peptide bond behind aspartate in one procaspase, the pro-domain is released and at the same time the part α and β separates from each other to form the active heterotetramer α 2 β 2 .
The cleavage itself proceeds by a mechanism similar to the catalysis of serine proteases (as a nucleophile, sulfur acts in place of oxygen). The cysteine residue and the histidine residue are close to each other in the active site of the enzyme. The rest of histidine receives a hydrone from the -SH group of cysteine and thus activates it. The negatively charged sulfur nucleophilically attacks the carbonyl peptide bonds of aspartate and subsequent amino acids of the cleaved protein, thereby binding the aspartate. Histidine moves the hydron to the amino group of the next amino acid, thereby self-cleaving the peptide bond and leaving the aspartate-terminated portion of the protein bound to the cysteine, while the portion beginning with the following amino acid separates. Subsequently, the bond between the cysteine sulfur and the aspartate carbon is hydrolyzed, and the protease is regenerated.
However, the activation of apical (initiating) caspases is more complicated, as adapter proteins are not specific proteases and therefore cannot cleave the initiating caspase in the same way that it activates the effector caspase. The most likely theory is the mechanism by which an adapter protein binds (e.g., to an apoptosome or similar structure) two initiating procaspases and alters their conformation to activate each other. However, certain inconsistencies still need to be explained to demonstrate this theory.
External activation pathway[edit | edit source]
The external pathway of apoptotic caspase cascade activation is most often described as the pathway through the Fas receptor and procaspase-8 activation. It is triggered by NK cells or activated T-lymphocytes that have Fas ligand in their membrane. Upon approaching a cell that is to undergo apoptosis, Fas ligand binds to its Fas receptor. It is a transmembrane protein which, when Fas-L binds, forms a complex of caspase-10, caspase-8 molecules and their adapter protein, which ensures the appropriate conformation of caspase-8, which reciprocally cleaves and thus activates. In a simplified scheme, caspase-8 activates procaspase-3 to caspase-3, which as an effector enzyme transmits the final signal to the onset of apoptosis (laminae, CAD and others).
Internal activation pathway[edit | edit source]
The intrinsic activation pathway is associated with cell damage. One of the mechanisms of induction of apoptosis after injury is the release of cytochrome c from the mitochondria. It travels to the cytosol and activates an adapter protein called Apaf-1, which activates procaspase-9 to caspase-9, which in turn activates effector caspases. This pathway is regulated at the level of cytochrome c binding and Apaf-1 proteins from the Bc-2 family: apoptosis is promoted by Bad (inactivates inhibitors), Bax and Bak (help release cytochrome c), while Bcl-2 and Bcl-X L inhibit it (by preventing release of cytochrome c). Another way of regulation are proteins from the IAP family ( inhibitor of apoptosis family ), which inhibit the activation of caspases, or directly their activity. This can be used, for example, by virusesto prevent the invaded cell from apoptosis.
The internal pathway can also trigger DNA damage. The signal is usually transferred via the p53 gene, which activates the transcription of genes that encode proteins that promote cytochrome cz mitochondria release .
Links[edit | edit source]
References[edit | edit source]
- VOET, Donald and Judith VOET. Biochemistry. 3rd edition. John Wiley & Sons Inc, 2004. 1616 pp. ISBN 0-471-19350-X .
- ↑ ALBERTS, Bruce, Alexander JOHNSON and Julian LEWIS, et al. Molecular Biology Of The Cell [online] . 4th edition. USA: Garland Science, 2002. Also available from < https://www.ncbi.nlm.nih.gov/books/NBK26873/ >. ISBN 0-8153-3218-1 .
- ↑ HUGO Gene Nomenclature Committee. Gene Nomenclature Committee [online]. © 2010. [feeling. 5.5.2010]. < https://www.genenames.org/ >.
- ↑ NAGATA, Shigekazu. Apoptotic DNA Fragmentation - minireview. Experimental Cell Research [online] . 2000, vol 256, pp. 12-18, also available from < http://www.sciencedirect.com/science/journal/00144827 >. ISSN 0014-4827.
- ↑ Advances in Experimental Medicine and Biology, Vol. 615.. Programmed Cell Death in Cancer Progression and Therapy, Caspase Mechanism [online] . XIV edition. Springer, 2008. 356 pp. Also available from < https://www.springer.com/gp/book/9781402065538 >. ISBN 978-1-4020-6553-8 .
Related articles[edit | edit source]
Recommended literature[edit | edit source]
- POP, Cristina and Guy S. SALVESEN. Human Caspases: Activation, Specificity, and Regulation. The Journal of Biological Chemistry [online] . 2009, vol. 284, pp. 21777-21781, also available from < http://www.jbc.org/content/284/33/21777.abstract >. ISSN 1083-351X.