Urine protein determination
Qualitative determination of protein in urine[edit | edit source]
Diagnostic strips are used to detect pathological proteinuria. In some laboratories, the strip test is combined with a sulfosalicylic acid test.
Diagnostic strips[edit | edit source]
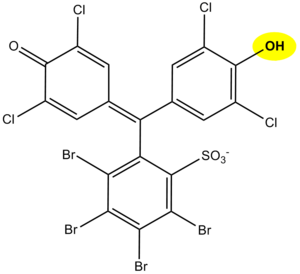
The principle of protein determination in urine using diagnostic strips is based on the so-called protein acid-base indicator error, e.g. tetrabromophenol blue, tetrabromophenolphthalein ethyl ester, or 3´,3´´,5´´,5´´-tetrachlorophenol-3,4,5,6-tetrabromophenolphthalein. Like any acid-base indicator, these substances change colour at a certain pH (they behave like weak acids, with the protonated form having a different colour than the dissociated form): at pH lower than 3.5 they are yellow, at higher pH they are green to blue. In addition to the indicator, the reaction zone of the test strip contains a buffer which maintains the pH between 3,0 and 3,5, so the indicator is yellow. If there are proteins in the sample, they bind to the indicator with their amino groups. However, this changes its properties – the transition region shifts towards the more acidic pH. This means that at the constant pH between 3.0 and 3.5, the indicator with the protein bound will be green, as if it were in a more alkaline environment (hence the protein indicator error). The intensity of the colour depends on the concentration of the protein, ranging from green to blue, and is assessed visually or instrumentally.
For strongly alkaline urines (pH above 8) or if the urine is very concentrated, the test may give false positive results (buffer depletion in the reaction zone). In these cases, we acidify the urine with a few drops of dilute acetic acid to pH 5-6 and repeat the test. False positives can also be caused by high concentrations of certain substances with amino groups (contamination of the collection vessel by certain disinfectants), which bind to the indicators in a similar way to proteins.
The disadvantage of the test strips is their different sensitivity to individual proteins. The strips react very well with albumin and indicate its presence in urine from 0.1 to 0.5 g/l. They show significantly lower sensitivity to globulins, glycoproteins, and Bence-Jones protein. These diagnostic strips cannot demonstrate the increase in albuminuria to values up to about 200 mg/l or the daily albumin loss of 30 to 300 mg/24 hours that accompanies the earlier stages of some nephropathies. Immunochemical methods such as special diagnostic strips based on immunochromatographic principles or immunoturbidimetry can be used to screen for an increase in albuminuria.
Sulfosalicylic acid test[edit | edit source]
The principle of the test is denaturation of the protein by sulfosalicylic acid, which results in opalescence to opacity.
The reaction is sensitive, showing 0.1-0.2 g/l of total proteinuria. Differences in the detection of individual proteins are not as pronounced as with diagnostic strips. Sulfosalicylic acid also precipitates globulins.
False-positive results are given by this test when certain X-ray contrast agents, penicillin, sulphonamides, salicylic acid, and antidiabetic drugs are excluded.
A semi-quantitative scale is used for evaluation:
| Finding | Evaluation | Approximate concentration of protein in g/l |
|---|---|---|
| Opalescence | trace amounts | 0,05–0,1 |
| Light haze (transparent, underlying text is readable) | + | 0,1–0,2 |
| Milky haze (opaque, without flakes) | ++ | 0,5–1,0 |
| Milky haze with flake formation | +++ | 2,0–5,0 |
| Flaky clot | ++++ | ≥ 5,0 |
Quantitative determination of protein in urine[edit | edit source]
Quantitative determination of protein in urine is methodologically quite difficult. In clinical-biochemical practice, methods are used for the determination of proteinuria, which can be divided into three groups according to the principle:
- methods based on denaturation followed by turbidimetric determination of the haze (e.g. methods with trichloroacetic acid or sulfosalicylic acid);
- colorimetric methods with or without prior denaturation of proteins (e.g. biuret reaction);
- methods based on binding of dyes to proteins (e.g., the method with Coomassie brilliant blue G 250 according to Bradford, with pyrogallol red, etc.).
Today, techniques that can be automated - turbidimetry and colour reaction with pyrogallol red - are preferred.
Determination of proteinuria using pyrogallol red[edit | edit source]
Pyrogallol red is used for protein quantification, e.g. for the determination of proteinuria. Pyrogallol red forms a pink complex with molybdenum with an absorption maximum at 470 nm. When the protein binds to this complex in an acidic environment, the colour deepens and the absorption maximum shifts to the 600 nm region. The absorbance of the resulting complex of reagent with protein is linearly dependent on the concentration of protein in the sample in the concentration range 0.06-2.0 g/l.
References[edit | edit source]
Related articles[edit | edit source]
References[edit | edit source]
- ↑ KRAML, Jiří, et al. Návody k praktickým cvičením z lékařské chemie a biochemie. 1. edition. Praha : Karolinum, 1991. 312 pp. pp. 243. ISBN 80-7066-453-3.