Monohybridism
A hybridization experiment, where the inheritance of one trait determined by one gene is monitored, is called monohybridism. Hybridization experiments belong among the basic methods of experimental genetics. The knowledge gained can subsequently be used in monitoring the inheritance of monogenically determined traits in the human population. Suppose that the gene encoding a monogenically determined trait is located on a autosome.The sign occurs in two distinct forms. It follows that there are two forms of this gene (alleles) in the population, namely the dominant allele (conventional signification A) and the recessive allele (conventional signification a). The hybridization experiment is based on the parental generation (cross) marked P, when the parents are different homozygotes for the given gene, i.e. they are pure lines. In our case, one of the parents is a dominant homozygote (AA), the other is a recessive homozygote (aa). When crossing, it does not matter what gender the two homozygotes are (this does not apply to genes located on sex chromosomes).
- Female x male: either AA x aa, or aa x AA
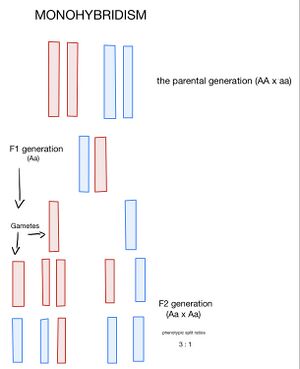
The bar chart (see image) shows a hybridization experiment where one gene per pair of autosomes is monitored (monogenic inheritance). The autosome carrying the dominant allele A is red, the autosome carrying the recessive allele a is blue. Individuals of the parental generation are all gametes with a haploid number of chromosomes with the corresponding genetic gear (A or a). Individuals of the first filial generation (F1) arise from the fusion of the gametes of the parents and will therefore all be Aa heterozygotes. The F1 generation is uniform. Both sexes form two types of gametes, each with a 50 % probability. The mutual crossing of females and males of the F1 generation (intercross) results in a second filial generation (F2). Genotype combination in the F2 generation can be deduced using a Punnett combination square (see bar chart). Genotypic split ratios in the F2 generation are 1 (AA) : 2 (Aa) : 1 (aa). Phenotypic split ratios are 3 : 1 assuming complete dominance of A over a , when only in the case of a recessive homozygote (aa) the influence of the dominant allele is not realized.