DNA Sequencing
Sequencing is a collective term for methods that allow describing the sequence of nucleotides in a certain section of DNA . These methods can be used to indirectly analyze the RNA sequence , if it is converted to DNA by reverse transcription . In practice, two methods based on DNA replication are used today - the older Sanger sequencing and the newer, in principle more complex next generation sequencing - Next Generation Sequencing .
Historical methods of sequencing
Older sequencing methods (unlike newer ones) do not use the principle of replication, but nucleic acid cleavage.
Before determining the nucleotide sequence , the nucleic acid is first cleaved with the aid of an endonuclease enzyme into shorter chains (oligonucleotides). By partially cleaving the oligonucleotide with e.g. phosphodiesterase from snake venom, a mixture of grafts differing by one nucleotide at the 3'-end can be obtained (e.g. from CCUAGCA a mixture containing CCUAGCA, CCUAGC, CCUAG, CCUA, CCU, CC is formed). These individual grafts are recognized by size using electrophoresis . The representation of bases must be determined in each electrophoretic fraction. This procedure was the principle of the first method of determining the sequence of nucleotides in RNA. Its lengthy nature is obvious, it was necessary to analyze individual short oligonucleotides in the described manner and then to sort them based on the analysis of at least two starting nucleic acid lysates obtained by two different endonucleases with different specificities.
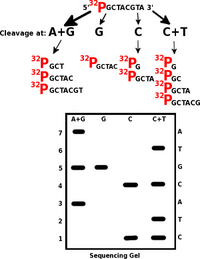
Maxam and Gilbert found a faster procedure when they used chemical cleavage of the polynucleotide at one of the four nucleotides to determine the primary structure of nucleic acids ( sequencing ) (guanine nucleotides can be cleaved with dimethyl sulfate, cytosine nucleotides with piperidine, etc.). The oligonucleotide is first labeled at the 5'-end with radioactive 32 P and its solution is divided into four equal samples. Each sample is cleaved preferentially at one of the four nucleotides. The chosen conditions allow mostly one cleavage in the molecule. So, for example, in the oligonucleotide 5' - 32 P-TACGTCGA, four different mixtures of grafts are obtained.
These fragments are then size-separated by polyacrylamide electrophoresis , and the fractions are detected by autoradiography or fluorescence. The sequence can be read directly from the electrophorogram. The evaluation proceeds from the shortest fragment and the type of lysate in which the graft is found (A, G, C or T) is recorded.
Sanger sequencing[edit | edit source]
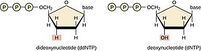
In 1977, Frederick Sanger created the sequencing method for which he received the Nobel Prize in 1980. The basis is modified DNA replication – standard deoxyribonucleotide triphosphates (dNTPs) are mixed with special dideoxyribonucleotide phosphates (ddNPTs) in the replication mixture. They are chemically similar to each other, but the missing OH group of ddNTP does not allow replication to continue.
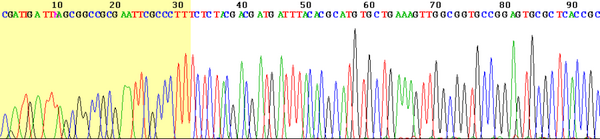
In the modern version, the reaction takes place as follows – the reaction mixture contains template (examined) DNA , DNA polymerase , sequencing primers and dNTPs (i.e. dATP, dGTP, dCTP, dTTP), i.e. all substrates of replication. Furthermore, the mixture contains fluorescently labeled ddNTPs . Together with buffer solutions, necessary ions and other stabilizers, this mixture of reagents for PCR DNA amplification is referred to as a mastermix. Here, the DNA polymerase replicates the examined DNA template molecule in the section bounded by the sequencing primers, for which it uses a mixture of dNTPs and ddNTPs. These nucleotides are incorporated into the strand based on complementarity to the template DNA just as in normal replication. Randomly incorporated ddNTPs, however, terminate the reaction with the formation of fragments of various lengths. By analysis using capillary electrophoresis , we then sort the individual fragments according to their length, so that the terminal nucleotides of the gradually lengthening fragments form a complete series if a sufficient number of reactions have taken place.
Fluorescence peaks marking the individual bases of the terminal ddNTPs will allow the resulting sequence to be read. Such data can be analyzed by a computer program. The first few bases are usually worthless - in the case of short fragments, non-specific binding occurs and little of the desired replication product is produced. It is therefore necessary to design primers sufficiently in front of the site that is the target region (target) for sequencing. The following figure shows the result.
The original method of 4 separate tubes
Before linking with capillary electrophoresis and fluorescent labeling, the reaction was divided into 4 separate tubes according to the original Sanger protocol. Only one of the ddNTPs was present in each, resulting in fragments ending in only one base. The products formed in all 4 tubes were then lined up next to each other on gel electrophoresis, allowing the resulting sequence to be read. The result is similar to the Maxam-Gilbert method based on DNA cleavage.
Example: In the left column of the picture we see fragments of different lengths ending with adenine, in the other columns with other bases. By lining up all the fragments next to each other, we can read the resulting DNA sequence. In the picture we can see that with standard writing from the 5' end to the 3' end we subtract the TACAG sequence...
Next Generation Sequencing[edit | edit source]
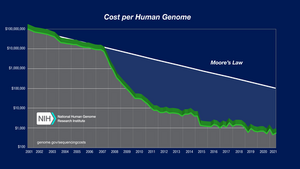
Next generation sequencing ( NGS ) is the name of modern sequencing methods that use bioinformatics methods to process large amounts of sequencing data and compare them with the reference genome . In NGS, thousands to millions of sequences are processed simultaneously, resulting in huge amounts of output data (hence sometimes also called massively parallel sequencing). Sequencing principles are different for individual manufacturers, as are sample preparation methods. One run of the sequencer is more expensive compared to the Sanger method and requires more complex equipment and reagents, but it will enable the sequencing of a large amount of source DNA in a short time and, with sufficient turnover, at a very low price compared to the Sanger method. With NGS, long sections of DNA (up to the entire genome ), multiple sections of DNA from one sample, one section in many repetitions to increase measurement accuracy, or many similar sections of DNA from different samples can be sequenced. Over the past 20 years, the cost of sequencing the human genome has dropped from millions of dollars to hundreds thanks to NGS.
There are very fundamental differences between individual NGS methods. Today, the most commonly used method is sequencing by synthesis , other principles include pyrosequencing, ion semiconductor sequencing or ligation sequencing. There are also differences in sample preparation depending on the procedure required.
Preparation of the sequencing library[edit | edit source]
Sequencing of the new generation is preceded by a relatively long pre-analytical phase, during which a solution prepared for sequencing on the given instrument is created, the so-called sequencing library :
- First (as with other sequencing methods) DNA must be isolated from the sample. Using various methods, we achieve the removal of cell membranes, proteins, carbohydrates, lipids or other substances, such as compounds used to fix the sample during its storage.
- After a similar process of RNA isolation, it is possible to use reverse transcriptase enzyme to convert RNA to DNA and sequence the bases of the transcriptome .
- In the case of some procedures, parts of the DNA that we want to sequence are amplified using PCR . First, however, the concentration of the input DNA and the integrity of the fragments are verified (that is, if the isolated DNA molecules are long enough and there are enough of them in the sample for analysis). Some of the less frequently used NGS methods do not need pre-amplification.
- Depending on the chosen sequencing method, the samples and sequencer reaction mixture must be adequately prepared. The two basic methods of library preparation are amplicon and enrichment sequencing:
- During amplicon sequencing , we use specifically defined primers for the given sections of DNA that we sequence. The sections are short and are sequenced in many repetitions. In this way, we can focus on specific changes in the sequence of a given gene in detail with high accuracy - this method is most often used for sequencing many shorter samples, for example when screening for tumor mutations in several patients.
- Enrichment sequencing works with much longer fragments, making it possible to more effectively examine longer sections of DNA, for example during whole-genome sequencing, resequencing or searching for new or unknown mutations in a wider area. In this method, labeled probes are used, only specific sections defined by these probes enter the amplification after purification on magnetic particles. This step reduces the formation of unwanted amplification products arising during amplicon sequencing by non-specific multiplication of short sequences when amplicons overlap.
- Depending on the method used, the affiliation of the sequence to the given sample is also marked in different ways - due to the fact that several samples are sequenced in parallel, for example , indexes are used , which are short oligonucleotides added to the beginning or end of the monitored sequence. Some of the sequencing methods also require the attachment of so-called adapter sequences, which enable fixation of the prepared DNA fragments to the given site intended for its sequencing.
Sequencing by synthesis and bioanalytics[edit | edit source]
Sequencing by synthesis is the most commonly used method of self-sequencing, in 2013 it covered about 90% of the market [1]. It is therefore presented here as the most common example of sample processing after the previously described preparation of the sequencing library.
Using the adapter sequences, the analyzed fragments are attached to the flow-cell sequencer (a glass plate with millions of fixed oligonucleotides complementary to the adapter sequences). Following other reactions, the DNA is replicated and the replicas are attached as additional templates to the flow-cell . During DNA sequencing, polymerases catalyze ongoing replication with fluorescently labeled dNTPs, so the instrument records different color signals for individual bases in a well-defined network of flow-cell binding sites . Schematically, this four-color image of the flow-cell at one moment is shown in the figure on the right. The instrument obtains hundreds of reads in both directions from one sample DNA fragment in a few reads.
The resulting "video" capturing millions of simultaneously scanned changing color points is analyzed by a computer program that assembles individual sequences, assigns them to patients/samples according to index sequences, evaluates the similarity of measurements of the same sections and the amount of deviations between measurements and compared to the reference genome. During the so-called alignment , the software evaluates according to the prescribed data, which sections scanned during sequencing will be compared with the reference sequence, and removes excess data (redundantly sequenced parts of DNA, non-specific replicates, primer sequences, etc.)
Video describing sequencing by synthesis[edit | edit source]
Other NGS methods[edit | edit source]
Different manufacturers use alternative next-generation sequencing methods. Pyrosequencing uses reversible termination of transcription with the help of several linked enzymatic reactions. This method was the first NGS method introduced to the market, but in order to be surpassed by other manufacturers, the production of new devices was gradually discontinued by the original manufacturer after 2013 [2].
Sequencing with the help of oligonucleotide ligation uses a labeled reaction in which the ligase enzyme attaches oligonucleotides with a known sequence to the primer based on complementarity to the template DNA. This method has the lowest error rate.
Other methods, such as sequencing on a semiconductor chip or nanopores, are rarely used.
The use of sequencing in medicine[edit | edit source]
Both Sanger sequencing and NGS have wide applications in multiple fields of medicine. This is preventive/predictive medicine – we use biomarkers of the risk of disease development, for example BRCA mutations ; diagnostic analyzes enabling the confirmation of the given disease or its extent ( Huntington's chorea ) as well as the examination of predictive or prognostic biomarkers that describe the severity of the given disease and its possible treatment. An example of these examinations is, for example, the evaluation of HER-2/neu receptor mutations in breast tumors or mutations of tyrosine kinase signaling cascades in colorectal cancer . Furthermore, sequencing is used in forensic medicine or microbiology.
Sanger sequencing is primarily used when small amounts of short samples need to be sequenced. However, when sequencing larger amounts of data, its advantages are in most cases outweighed by the capabilities of NGS.
Links[edit | edit source]
Related articles[edit | edit source]
- The structure of nucleic acids
- Basic components of nucleic acids
- Primary structure of nucleic acids
- Cleavage of nucleic acid by hydrolysis
- Secondary structure of DNA
- Denaturation of nucleic acids, molecular hybridization
- Secondary structure of RNA
- Topology of DNA
- Interaction of DNA with proteins
- Bacterial chromosome
- Eukaryotic chromosomes
- Mitochondrial DNA
Reference[edit | edit source]
- ↑ REIFOVÁ, Radka. Sekvenování Nové Generace : Přednáška předmětu Genetické metody v zoologii [online]. Přírodovědecká fakulta Univerzity Karlovy, ©2013. [cit. 2021-06-14]. <http://web.natur.cuni.cz/zoologie/biodiversity/prednasky/GenetickeMetodyVZoologii/Prednasky_2013/NextGenerationSequencing_2013.pdf>.
- ↑ GenomeWeb. Roche Shutting Down 454 Sequencing Business [online]. The last revision 16-03-2013, [cit. 2021-06-14]. <https://www.genomeweb.com/sequencing/roche-shutting-down-454-sequencing-business>.
External links[edit | edit source]
- Illumina. NGS for beginners [online]. [cit. 2021-05-26]. <https://www.illumina.com/science/technology/next-generation-sequencing/beginners.html>.
- NOVOTNÁ, Marcela. Princip NGS metody [online]. ©2018. [cit. 2021-07-04]. <https://www.vuab.cz/princip-ngs-metody/>.
References[edit | edit source]
- BEHJATI, Sam – TARPEY, Patrick S. What is next generation sequencing?. Arch Dis Child Educ Pract Ed [online]. 2013, vol. 98, no. 6, p. 236-8, Available from <https://doi.org/10.1136/archdischild-2013-304340>. ISSN 1743-0585 (print), 1743-0593.
- Přírodovědecká fakulta Jihočeské univerzity v Českých Budějovicích. Sekvenování, genomika (přednáška z předmětu Základy moderní biologie) [online]. [cit. 2021-05-24]. <https://www.prf.jcu.cz/zmb/menu/sekvenovani-genomika.html>.
- ŠTÍPEK, Stanislav. Stručná biochemie. 1. edition. Medprint, 1998. pp. 13–14. ISBN 80-902036-2-0.